New Vistas in the Management of Treatment-Refractory Psychiatric Disorders: Genomics and Personalized Medicine
Abstract
To date, most therapies of treatment-refractory psychiatric disorders involve modifications of existing pharmacological treatments and/or cognitive therapies. However, many of these strategies are unable to bring these disorders into remission; therefore, a substantial number of patients with severe psychiatric disorders continue to have marked functional limitations. Newer therapies involve novel pharmacological approaches and novel psychotherapies as well as altering neuronal circuitry via techniques such as deep brain or transcranial magnetic stimulation; however, these newer therapies are also not completely efficacious. With the 10-year anniversary of the sequencing of the human genome, genetic analysis emerges as an important approach in assessing therapeutic response. In this review, the authors will examine the current knowledge of gene and environmental interactions in treatment response for a subset of psychiatric diseases.
DEFINING “PERSONALIZED MEDICINE”
The main theory behind personalized medicine is that each individual is unique and differences among patients should be considered when the proper therapies are determined. At its root, personalized medicine involves the systematic use of information obtained from patients to dictate the type and course of treatment. This information can take on many forms, including clinical diagnostics, environmental factors, epigenetic modifications, and biomarker changes, as well as maps of the relationship between genomics and therapeutic outcomes. In this review, we will focus on personalized medicine as it relates to individual differences in the genome.
DEFINING “TREATMENT REFRACTORY”
Understanding which patients do not respond to treatment is superficially simple; however, several considerations are important when a clinician is determining whether a patient is truly nonresponsive to treatment. These considerations fall into three main categories: 1) diagnostic issues, 2) treatment effects, and 3) environmental interactions. Accurate diagnosis is crucial to determining drug and/or therapy effectiveness because patients can present with comorbidities as well as have changes in the course of their disease, both of which create difficulties in determining whether disorders are truly refractory. Treatment effects can occur when there has been a history of polypharmacy as well as issues related to dosages and treatment length, both of which will confound the assessment of whether patients are truly nonresponsive. Environmental effects can also hamper the indication of lack of efficacy because socioeconomic factors can contribute to differences in compliance and/or adherence, as well as modify the relationship between drugs and treatments via external factors that may not be considered in most assessments (e.g., trauma outcomes).
GENETIC INFLUENCES ON PSYCHIATRIC ILLNESS AND DRUG RESPONSE: DIRECT EFFECTS
To date there have been three large National Institutes of Health-funded studies focused on psychiatric illness and treatment response. First, the Sequenced Treatment Alternatives to Relieve Depression (STAR*D) trial followed patients with nonpsychotic major depression for up to 6 years through a sequence of alternative treatment regimens (1). Second, in the Clinical Antipsychotic Trial of Intervention Effectiveness (CATIE), patients with schizophrenia were treated with several leading antipsychotics and followed for up to 18 months (2). Finally, the Systematic Treatment Enhancement Program for Bipolar Disorder (STEP-BD) was focused on patients with bipolar disorder who were treated with mood stabilizers and antidepressants following expert consensus guidelines (3). In each of these trials, only a relatively small percentage of patients achieved symptom remission, particularly after their first treatment regimen, and many patients within each study discontinued treatment either due to drug side effects or lack of efficacy. Currently, there are on-going examinations of the genetics of susceptibility and treatment response in each of these cohorts, as well as separate studies examining candidate genes and risk for disease.
Major depression
Candidate genes.
Meta-analysis of five twin studies has demonstrated that the heritability of major depressive disorder (MDD) is estimated at 37%, suggesting that there are both environmental and genetic factors influencing risk for MDD (4). One of the most well-studied neural systems in MDD is the serotonergic pathway. The first reported associations were with a variable number tandem repeats (VNTR) variant and an insertion/deletion polymorphism near the serotonin transporter gene (5, 6). The 5-HTT-linked insertion/deletion polymorphic region (5-HTTLPR) maps 1.4 kilobases upstream of the translational start site. The short variant has been shown to restrict transcriptional activity of the serotonin transporter (5-HTT) promoter, leading to low functional expression of the 5-HTT; thus, the risk allele is most likely acting via changing expression of the 5-HTT transcript, resulting in less presynaptic uptake of serotonin (6, 7). Further work has focused on environmental influences modifying the relationship between 5-HTTLPR and risk. Caspi et al. (8) demonstrated that individuals with one or two copies of the short allele of the 5-HTTLPR promoter polymorphism exhibited more depressive symptoms, diagnosable depression, and suicidality in relation to stressful life events in children (i.e., abuse and neglect) than individuals homozygous for the long allele. Grabe et al. (9) found significant interactions between genotype, unemployment, and chronic diseases in females but not in males, further validating the modifying response of trauma and 5-HTT variation. This result further implicated the short allele of the 5-HTTLPR promoter polymorphism in vulnerability to social stressors and chronic disease.
In terms of pharmacogenetic response, serotonin reuptake inhibitors (SSRIs) act via inhibition of presynaptic serotonin reuptake; therefore, it makes sense to evaluate 5-HTT genotypes and SSRI response. A recent study comparing both the 5-HTTLPR promoter polymorphism and an intronic VNTR in 5-HTT in a group of SSRI nonresponders with a referent sample found that women with the short allele of 5-HTTLPR were less likely to respond to SSRI treatment, whereas the different alleles of the intronic polymorphism had no affect on response (10). This result was in contrast with results from a study on 241 Korean inpatients examining 5-HTT genotypes and fluoxetine or sertraline response (11). In the Korean patients, the short allele of 5-HTTLPR was associated with a positive response to treatment. This result reflects different outcomes due to either ethnicity effects or differences in study design. Another possibility is that the disparate results are indicative of 5-HTT being a false-positive response, which is supported by the negative findings from the STAR*D trial (12). Further work needs to be performed to assess 5-HTT and SSRI response.
Additional work on serotonergic pathways and MDD has implicated the serotonin 5-HT2A receptor (HTR2A) and more recently the serotonin 5-HT3A receptor (HTR3A). Whereas most of the studies of serotonergic pathways have focused on schizophrenia and psychosis (see next section), McMahon et al. (13) found an association between variation in intron 2 of HTR2A and response to citalopram in patients with MDD. Citalopram down-regulates the serotonin 2A receptor, which is encoded by the HTR2A gene. Another important factor in this association was that the allele involved in citalopram response was more frequent in Caucasian subjects, and treatment effectiveness is reduced in African American subjects. Therefore, the HTR2A allele might account for some of the ethnic differences in treatment response in MDD. Another study examining the −42C>T of HTR3A demonstrated that CC carriers had frontolimbic gray matter alterations, which were potentiated by early life stress and these changes could increase risk for MDD (14).
Besides serotonin, the other major pathway that is crucial in the causality as well as the treatment of depression is the hypothalamic-pituitary-adrenal (HPA) axis. The HPA axis is a major part of the neuroendocrine system that controls the reaction to stress as well as many other bodily functions. There is a complex interaction between the different HPA axis thyroid hormones in MDD (15), and recent evidence suggests that allelic variation is a factor in modifying both risk and drug response.
The first member of the HPA axis that was implicated in MDD was the FK506-binding protein 5 gene (FKBP5). Binder et al. (16) genotyped variants in eight genes involved in the HPA axis (16) and found that variation in FKBP5 was significantly associated with increased recurrence of depressive episodes as well as faster response to antidepressant drug treatment. The mapped risk variants were also associated with increased intracellular FKBP5 protein expression, which triggers adaptive changes in the glucocorticoid receptor and, thereby, modifies HPA axis regulation.
The second HPA axis gene to be associated with MDD was the corticotropin-releasing hormone receptor 1 (CRHR1) gene. Corticotropin-releasing hormone is the principal neuroregulator of the HPA axis and plays an important role in coordinating the endocrine, autonomic, and behavioral responses to stress and immune challenge. The first associations, which mapped three variants, one of which was associated with disease risk, were found in Chinese cohorts (17). That work was extended both within the same cohorts (18) as well as in another cohort of Mexican Americans (19), showing haplotype affects with treatment response for fluoxetine or desipramine. More recent work has now included CRHR1 gene × trauma interactions on MDD risk, showing that certain alleles act synergistically with child abuse to increase risk (20).
More recently, polymorphisms within the corticotropin-releasing hormone-binding protein (CRHBP) gene were assessed with respect to response to citalopram using the STAR*D samples. CRHBP is another member of the HPA axis and is a high-affinity binding protein that inactivates corticotropin-releasing factor. One variant (rs10473984) showed a significant association with both remission (and reduction in depressive symptoms in response to citalopram) (21).
Along with serotonin, there is some evidence to suggest that norepinephrine pathways are involved in MDD and treatment response. Two studies have examined drug response and variation in the norepinephrine transporter gene (SLC6A2). The first study (22) found that the T allele of the SLC6A2 T182C polymorphism was associated with an improved response to milnacipran. The second study (23) found that variants in the norepinephrine transporter gene (SLC6A2) predicted response to nortriptyline.
Genome-wide association studies.
Four genome-wide association studies of antidepressant response have been published (24, 25, 27, 28). This first study involved assessment of individuals from the STAR*D sample who developed treatment-emergent suicidal ideation with citalopram (24). After the analysis of ∼110,000 single nucleotide polymorphism (SNPs), a single marker mapping to the Papilin (PAPLN) gene was significant after correction for multiple testing. Little is known about PAPLN other than that it is a proteoglycan-like sulfated glycoprotein; it is unclear how this gene influences suicidal outcomes in citalopram treatment.
The second study again used the STAR*D cohort to examine the efficacy response to citalopram considering alleles at ∼430,000 variants (25). Although no variants reached genome-wide significance after correction for multiple testing, three loci showed suggestive evidence of being involved in treatment response. Variants near the ubiquitin protein ligase E3C (UBE3C), bone morphogenetic protein 7 (BMP7), and retinoic acid-binding receptor α (RORA). Although it is unclear how these novel genes might be involved in MDD, it is interesting to note that RORA might be involved in pathways integral to circadian rhythms in mammalian systems (26).
The third study used cohorts collected in the Munich, Germany, Antidepressant Response Signature (MARS) project that examined drug efficacy after antidepressants were prescribed according to doctor choice. The authors report that they mapped an aggregate series of markers that predict low versus high response to various drug treatments (27). This finding suggests that treatment response may be multifactorial and under the control of a number of additive genetic loci instead of a limited number with large effects.
The fourth genome-wide association study used samples from the Genome-Based Therapeutic Drugs for Depression (GENDEP) study, which is a partially randomized open-label pharmacogenetic trial (28). In this study, two intergenic regions containing copy number variants on chromosomes 1 and 10 were associated with the outcome of treatment with escitalopram or nortriptyline at suggestive levels of significance. Drug response to nortriptyline was best predicted by variation in the uronyl 2-sulfotransferase gene. Response to escitalopram was best predicted by a marker in the interleukin-11 (IL11) gene.
Schizophrenia
Candidate genes.
Concordance rates for schizophrenic monozygotic twins average ∼46%, even when they are reared in different families, whereas the concordance rates for dizygotic twins averaged only 14%, suggesting that like MDD, schizophrenia (SCZ) has both genetic and environmental components (29). Similar to MDD, serotonergic pathways have also been implicated in the pathophysiology of SCZ and efficacy of antipsychotics. Second generation (atypical) antipsychotics in particular display high affinity for 5-HT2 receptors, and this affinity is thought to mediate their action in part (30, 31). However, the specific serotonin receptors involved in SCZ seem to be quite different from those for MDD. Specifically, the HTTLPR polymorphism does not appear to be involved in SCZ risk (32, 33). Much work has focused on the HTR2A gene, and a meta-analysis demonstrated that a synonymous variant (T102C) in HTR2A is associated with disease risk (34); however, previous work suggested that perhaps it is not the T102C allele but a promoter polymorphism that is associated with risk (35). Further work has now shown that serotonergic systems might also be involved in drug response. In a study of risperidone response, patients who were HTR2A T/T homozygotes for the T102C allele showed less clinical improvement than did those with T/C or C/C genotypes (36). Mössner et al. (37) studied the serotonin 1A receptor (HTR1A), which mediates inhibitory neurotransmission. They assessed whether different alleles were modifying the response in terms of negative symptoms to risperidone or haloperidol. They found that carriers of the C allele were more likely to respond to treatment than the G allele. This was further confirmed in a study in Chinese patients (38). Another polymorphism (−759C/T) in the promoter of the HTR2C gene has also been associated with improvements in negative symptoms after risperidone or chlorpromazine treatment (39), Souza et al. (40) mapped variants in the serotonin 3 receptor (HTR3A and HTR3B) and showed that these were associated with response to clozapine.
Beyond the serotonergic pathways, there has been an extensive study of dopaminergic pathways and processing in SCZ. Dysregulation of the dopaminergic system was among the first and remains the preeminent pathological finding in schizophrenia, and dopamine D2 receptor blockade is the essential pharmacological feature of all antipsychotics. One of the most studied genes in terms of dopamine response is catechol-O-methyltransferase (COMT), which is a the major degradative pathway of catecholamines. The discovery of deletions that map to the same chromosomal region as COMT (22q11) in velo-cardio-facial syndrome (VCFS) was one of the first clues that COMT might be involved in SCZ. Both VCFS and SCZ have psychosis as a primary symptom, suggesting common mechanistic pathways (41). A common functional polymorphism (Val108/158Met) has been shown to be associated with SCZ as well as a fourfold variation in COMT activity and dopamine catabolism (42). Bray et al. (43) looked at the Val108/158Met variant and two additional coding variants within COMT and disease risk for SCZ (43). They found that SCZ risk was associated with a haplotype that resulted in a lower expression of the COMT transcript. This association was further validated in a study of 38 world populations (44) as well as a cohort from Korea (45). In terms of treatment response, COMT Met homozygotes with low enzyme activity have been shown to have more than 10 times higher risk of being a nonresponder than responder to treatment with typical neuroleptics (46). An association was also found in populations from south India (47) and China (48), looking at the responsiveness to risperidone treatment.
Another system implicated in drug response in SCZ is the N-methyl-d-aspartate (NMDA)/glutamate transmitter system. It has been proposed that hypofunction of this system might be crucial for the etiology and treatment of SCZ (49). Interestingly, one of the more recent genes implicated in SCZ seems to act through the NMDA pathway. Disrupted in schizophrenia 1 (DISC1) is unusual in that it was mapped as a SCZ gene via molecular cytogenetics as opposed to the more typical linkage or association mapping. Millar et al. (50) mapped two genes (DISC1 and DISC2) in a large Scottish family that had a balanced translocation between chromosomes 1 and 11, which interrupted DISC1 and DISC2 (antisense to DISC1) on chromosome 1. Of 77 family members with mental illness, 34 had the translocation, whereas none of the 38 members who did not have the translocation were affected. Sawa et al. (51) showed that DISC1 helps to regulate the plasticity of dendritic spines in response to NMDA receptor activation and thus might be crucial for neuronal plasticity in response to NMDA. In terms of pharmacogenetic response in SCZ, patients with DISC1 might represent a new subtype of SCZ that is typically resistant to treatment. In a Canadian cohort, a missense mutation in DISC1 was associated with an ultra-resistant phenotype characterized as patients who experience no clinical, social, and/or occupational remission despite treatment with clozapine and at least two periods of treatment with distinct conventional or atypical antipsychotic drugs (52).
Genome-wide association studies.
Three genome-wide association studies of antipsychotic treatment response have been reported in the literature (53–55). The first study (53) was derived from the CATIE trial in which patients with SCZ were randomly assigned to treatment with either a second-generation (olanzapine, quetiapine, risperidone, or ziprasidone) or a first-generation (perphenazine) antipsychotic. This study tested for genome-wide predictors of efficacy among 738 patients genotyped using the Affymetrix 500K genotyping platform supplemented with a custom 164K chip to improve genome-wide coverage. Efficacy was measured by changes over time in positive and negative symptom scores. Their top result mapped to an intergenic region on chromosome 4p15. Two other findings were close to their prespecified threshold for genome-wide significance mapped to the ankyrin repeat and sterile alpha motif domain-containing protein 1B (ANKS1B) gene and the contactin-associated protein-like 5 (CNTNAP5) gene, which were found to mediate negative symptom response to olanzapine and risperidone, respectively. The next genome-wide association study (54) used a phase 3 randomized trial of iloperidone, an investigational drug for the treatment of SCZ that has been in development for more than two decades. In this study ∼330,000 variants were assessed in 407 patients. The outcome was change from baseline to last scheduled observation in positive and negative total symptom scores. Three complementary analyses were performed, and six loci were identified with consistent findings across these analyses. The authors found results with the neuronal PAS domain protein 3 gene (NPAS3) as well as with five additional loci. The final study (55) took a genomic convergence approach to examine response to risperidone. These authors collapsed results from a 100,000-variant genome screen of risperidone response, with a transcriptome study of differential expression to risperidone treatment in mouse frontal cortex. They found 14 genes that overlapped between screens, which they carried into a second stage of replication looking at a case-control series, in which one gene (PDE7B) survived multiple testing correction. Care needs to be taken in interpreting these results because mouse brain expression is known to not fully map to pathways relevant in human cortex (56).
Adverse effects.
Another important consideration in the assessment of treatment response in SCZ is that many of the antipsychotic drugs have considerable side effects that are severe enough to alter compliance and treatment course. These include tardive dyskinesia (TD), weight gain, and glucose and lipid abnormalities. Many of these side effects have been examined to map the genetic profiles that render patients susceptible to these side effects.
Probably the best-studied adverse effect is TD, which sometimes occurs after a long-term course of treatment with first-generation antipsychotics such as haloperidol. TD is a persistent extrapyramidal syndrome that is characterized by involuntary movements in the tongue, lips, or jaw as well as facial grimacing, movements of arms, legs, fingers, and toes. Two genome-wide screens using the CATIE cohorts studied DNA variation and risk for TD (57, 58). The more inclusive study (58) examined symptoms of parkinsonism, akathisia, and abnormal involuntary movements. Three findings met genome-wide significance in novel regions that have not been previously implicated in the pharmacogenetics of extrapyramidal symptoms. Two were located in an intergenic region on chromosome 11q24, and the other was in ZNF202, which is a transcriptional repressor controlling PLP1, a major component of myelin. Two other studies in Japanese cohorts mapped additional regions involved in the development of TD (59, 60). The first study (59) found associations with eight genes involved in γ-aminobutyric acid signaling; however, the sample size was limited, and there was no replication. The second study (60) used the same cohort; however, the authors replicated their results using both an additional series as well as a knockdown animal model. They found an association with TD and the heparan sulfate proteoglycan 2, perlecan (HSPG2) gene that replicated in a second series. In heterozygous HSPG2 knockout mice, there was a reduction in vacuous chewing movements induced by 7-week injection of haloperidol-reserpine compared with that in wild-type littermates, showing that perturbing HSPG2 might protect against the development of TD-like symptoms.
BIPOLAR DISORDER
Candidate genes.
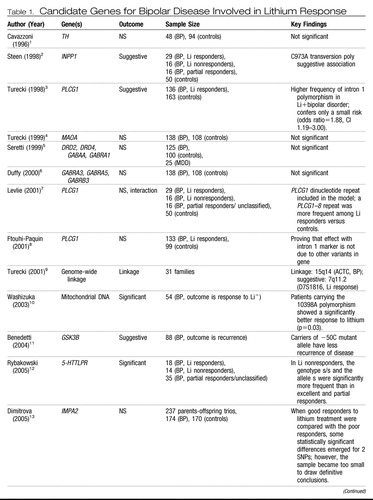
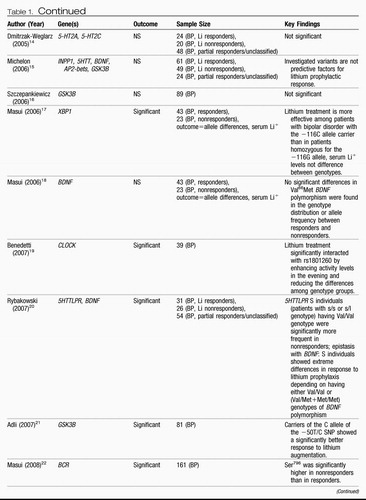
Twin and adoption studies have reported concordance rates of ∼38%–43% for monozygotic bipolar twins compared with a rate of ∼4%–5% for dizygotic twins, demonstrating that there is a very significant genetic component to the development of bipolar disease (61–63). The leading treatments for bipolar disorder (BPD) include lithium and the anticonvulsants, such as valproate, carbamazepine, and lamotrigine as well as atypical antipsychotics. There is a paucity of candidate gene pharmacogenetic studies of treatment response in BPD, with most of the studies being focused on lithium response. Table 1 summarizes the results from those screens.
 |
 |
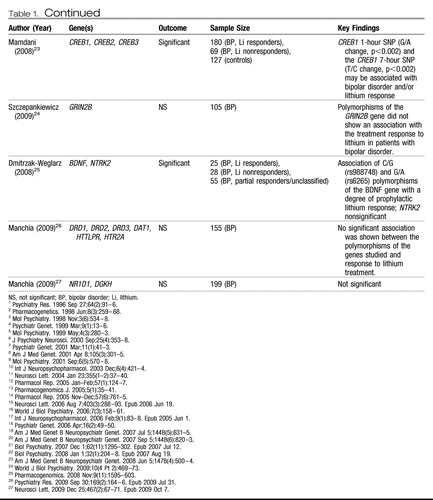 |
Table 1. Candidate Genes for Bipolar Disease Involved in Lithium Response
Genome-wide association studies.
Although there now have been several genome-wide screens mapping association or linkage and BPD risk, there are few pharmacogenetic genome-wide studies to date. One genome-wide screen on lithium response in BPD has been reported (64). This study used samples from the STEP-BD cohorts and prospectively examined whether there was differential recurrence comparing individuals receiving lithium alone or receiving lithium in combination with other psychotropics. Results from assessing 1.4 million markers were then replicated in an additional cohort from the United Kingdom. None of the SNPs tested in the STEP-BD cohort met genome-wide significant criteria for association. A total of 140 SNPs were carried forward for replication, however, and 9 were significant in the British sample at p<0.05. Of these, five results had the same direction of effect as in the initial STEP-BD screen and three displayed associations that were specific to lithium treatment. These variants mapped to an ionotropic glutamate receptor (GRIA2), a cell surface glycoprotein (SDC), and tenascin M4 (OZD), which is highly expressed in the nervous system and has been implicated in intracellular signaling during development and regulation of transcription (65).
GENETIC INFLUENCES ON PSYCHIATRIC ILLNESS AND DRUG RESPONSE: INDIRECT EFFECTS
Another important area for consideration in determining the relationship between genes and drug response in psychiatric disorders is the contribution of genetic polymorphism in drug-metabolizing genes. Unlike the genes that have been mapped for disease risk, genes that affect the pharmacodynamics of drug response are for the most part both highly penetrant and monogenetic.
Most medication is metabolized by one of the cytochrome P450 (CYP450) family of enzymes. The CYP450 isoenzyme CYP2D6 is responsible for metabolizing approximately 25% of drugs, including antidepressants (66). More than 90 genetic variants have been identified in CYP2D6 (67). These variants define four different groups based on enzyme activity: 1) poor metabolizers (PMs), 2) intermediate metabolizers (IMs), 3) extensive metabolizers (EMs), and 4) ultrarapid metabolizers (UMs). CYP2C19 is also polymorphic with two main phenotypic groups: EMs and the rarer PMs (68). CYP2D6 and CYP2C19 along with another CYP450 enzyme, CYP2C9, metabolize virtually all SSRIs (69). Antipsychotic drug metabolism is also largely determined by variants in CYP2D6 along with additional polymorphisms in CYP1A2, CYP3A4, and CYP3A5 (70). The generally accepted view has been that CYP450-related PMs will have an increased risk of side effects from antidepressants, whereas UMs and to a lesser extent IMs are less likely to show positive response to treatment. Based on this hypothesis, the AmpliChip CYP450 Test (71) was developed by Roche and approved by the U.S. Food and Drug Administration. The AmpliChip uses microarray technology from Affymetrix to type variants in CYP2D6 and CYP2C19, including 29 polymorphisms and mutations of the CYP2D6 gene and two polymorphisms of the CYP2C19 gene. The test includes software with an algorithm to predict CYP2D6 and CYP2C19 phenotypes (i.e., PM, IM, EM, and UM) based on the alleles identified.
The Centers for Disease Control and Prevention commissioned an independent panel to examine the use of CYP450 genotyping when SSRI antidepressants are prescribed (72). In their review, the independent panel determined that there was strong evidence for the analytic validity of CYP450 genotyping but only marginal evidence for its clinical validity and almost no evidence for its clinical use (73). Additional studies found that although the reliability of the AmpliChip was excellent (74), possible difficulties include implementation in real-world settings (75) as well as issues with differences in CYP450 allele frequencies due to ethnic background (76).
CONCLUSIONS
In this review, we have focused on mood disorders and schizophrenia and how genetics plays a role in treatment response. We have detailed the pharmacogenetics of individual syndromes, thus broadening the consideration of treatment efficacy prediction to other markers beyond imaging. Personalized medicine can and will be applied to other modalities beyond pharmacotherapy, including but not limited to psychotherapies (e.g., cognitive behavior therapy), and various nerve stimulation procedures (e.g., transcranial magnetic stimulation, vagal nerve stimulation, and deep brain stimulation). Moreover, other biological predictors of therapeutic response are under intense scrutiny, including EEG measures, transcript expression, protein expression, metabolites, and methylation patterns. Functional brain imaging ranging from functional magnetic resonance imaging to positron emission tomography and magnetic resonance spectroscopy are also active areas of investigation.
1 Rush AJ, Fava M, Wisniewski SR, Lavori PW, Trivedi MH, Sackeim HA, Thase ME, Nierenberg AA, Quitkin FM, Kashner TM, Kupfer DJ, Rosenbaum JF, Alpert J, Stewart JW, McGrath PJ, Biggs MM, Shores-Wilson K, Lebowitz BD, Ritz L, Niederehe G, STAR*D Investigators Group: Sequenced treatment alternatives to relieve depression (STAR*D): rationale and design. Control Clin Trials 2004; 25:119–142Crossref, Google Scholar
2 Stroup TS, McEvoy JP, Swartz MS, Byerly MJ, Glick ID, Canive JM, McGee MF, Simpson GM, Stevens MC, Lieberman JA: The National Institute of Mental Health Clinical Antipsychotic Trials of Intervention Effectiveness (CATIE) project: schizophrenia trial design and protocol development. Schizophr Bull 2003; 29:15–31Crossref, Google Scholar
3 Thase ME, Bhargava M, Sachs GS: Treatment of bipolar depression: current status, continued challenges, and the STEP-BD approach. Psychiatr Clin North Am 2003; 26:495–518Crossref, Google Scholar
4 Sullivan PF, Neale MC, Kendler KS: Genetic epidemiology of major depression: review and meta-analysis. Am J Psychiatry 2000; 157:1552–1562Crossref, Google Scholar
5 Ogilvie AD, Battersby S, Bubb VJ, Fink G, Harmar AJ, Goodwim GM, Smith CA: Polymorphism in serotonin transporter gene associated with susceptibility to major depression. Lancet 1996; 347:731–733Crossref, Google Scholar
6 Collier DA, Stöber G, Li T, Heils A, Catalano M, Di Bella D, Arranz MJ, Murray RM, Vallada HP, Bengel D, Müller CR, Roberts GW, Smeraldi E, Kirov G, Sham P, Lesch KP: A novel functional polymorphism within the promoter of the serotonin transporter gene: possible role in susceptibility to affective disorders. Mol Psychiatry 1996; 1:453–460Google Scholar
7 Bradley SL, Dodelzon K, Sandhu HK, Philibert RA: Relationship of serotonin transporter gene polymorphisms and haplotypes to mRNA transcription. Am J Med Genet B Neuropsychiatr Genet 2005; 136B:58–61Crossref, Google Scholar
8 Caspi A, Sugden K, Moffitt TE, Taylor A, Craig IW, Harrington H, McClay J, Mill J, Martin J, Braithwaite A, Poulton R: Influence of life stress on depression: moderation by a polymorphism in the 5-HTT gene. Science 2003; 301:386–389Crossref, Google Scholar
9 Grabe HJ, Lange M, Wolff B, Völzke H, Lucht M, Freyberger HJ, John U, Cascorbi I: Mental and physical distress is modulated by a polymorphism in the 5-HT transporter gene interacting with social stressors and chronic disease burden. Mol Psychiatry 2005; 10:220–224Crossref, Google Scholar
10 Smits KM, Smits LJ, Peeters FP, Schouten JS, Janssen RG, Smeets HJ, van Os J, Prins MH: The influence of 5-HTTLPR and STin2 polymorphisms in the serotonin transporter gene on treatment effect of selective serotonin reuptake inhibitors in depressive patients. Psychiatr Genet 2008; 18:184–190Crossref, Google Scholar
11 Kim H, Lim SW, Kim S, Kim JW, Chang YH, Carroll BJ, Kim DK: Monoamine transporter gene polymorphisms and antidepressant response in Koreans with late-life depression. JAMA 2006; 296:1609–1618Crossref, Google Scholar
12 Kraft JB, Peters EJ, Slager SL, Jenkins GD, Reinalda MS, McGrath PJ, Hamilton SP: Analysis of association between the serotonin transporter and antidepressant response in a large clinical sample. Biol Psychiatry 2007; 61:734–742Crossref, Google Scholar
13 McMahon FJ, Buervenich S, Charney D, Lipsky R, Rush AJ, Wilson AF, Sorant AJ, Papanicolaou GJ, Laje G, Fava M, Trivedi MH, Wisniewski SR, Manji H: Variation in the gene encoding the serotonin 2A receptor is associated with outcome of antidepressant treatment. Am J Hum Genet 2006; 78:804–814Crossref, Google Scholar
14 Gatt JM, Williams LM, Schofield PR, Dobson-Stone C, Paul RH, Grieve SM, Clark CR, Gordon E, Nemeroff CB: Impact of the HTR3A gene with early life trauma on emotional brain networks and depressed mood. Depress Anxiety 2010; 27:752–759Crossref, Google Scholar
15 Nemeroff CB: Clinical significance of psychoneuroendocrinology in psychiatry: focus on the thyroid and adrenal. J Clin Psychiatry 1989; 50(suppl):13–20; discussion 21–22Google Scholar
16 Binder EB, Salyakina D, Lichtner P, Wochnik GM, Ising M, Pütz B, Papiol S, Seaman S, Lucae S, Kohli MA, Nickel T, Künzel HE, Fuchs B, Majer M, Pfennig A, Kern N, Brunner J, Modell S, Baghai T, Deiml T, Zill P, Bondy B, Rupprecht R, Messer T, Köhnlein O, Dabitz H, Brückl T, Müller N, Pfister H, Lieb R, Mueller JC, Lõhmussaar E, Strom TM, Bettecken T, Meitinger T, Uhr M, Rein T, Holsboer F, Muller-Myhsok B: Polymorphisms in FKBP5 are associated with increased recurrence of depressive episodes and rapid response to antidepressant treatment. Nat Genet 2004; 36:1319–1325Crossref, Google Scholar
17 Liu Z, Zhu F, Wang G, Xiao Z, Wang H, Tang J, Wang X, Qiu D, Liu W, Cao Z, Li W: Association of corticotropin-releasing hormone receptor1 gene SNP and haplotype with major depression. Neurosci Lett 2006; 404:358–362Crossref, Google Scholar
18 Liu Z, Zhu F, Wang G, Xiao Z, Tang J, Liu W, Wang H, Liu H, Wang X, Wu Y, Cao Z, Li W: Association study of corticotropin-releasing hormone receptor1 gene polymorphisms and antidepressant response in major depressive disorders. Neurosci Lett 2007; 414:155–158Crossref, Google Scholar
19 Licinio J, O'Kirwan F, Irizarry K, Merriman B, Thakur S, Jepson R, Lake S, Tantisira KG, Weiss ST, Wong ML: Association of a corticotropin-releasing hormone receptor 1 haplotype and antidepressant treatment response in Mexican-Americans. Mol Psychiatry 2004; 9:1075–1082Crossref, Google Scholar
20 Bradley RG, Binder EB, Epstein MP, Tang Y, Nair HP, Liu W, Gillespie CF, Berg T, Evces M, Newport DJ, Stowe ZN, Heim CM, Nemeroff CB, Schwartz A, Cubells JF, Ressler KJ: Influence of child abuse on adult depression: moderation by the corticotropin-releasing hormone receptor gene. Arch Gen Psychiatry 2008; 65:190–200Crossref, Google Scholar
21 Binder EB, Owens MJ, Liu W, Deveau TC, Rush AJ, Trivedi MH, Fava M, Bradley B, Ressler KJ, Nemeroff CB: Association of polymorphisms in genes regulating the corticotropin-releasing factor system with antidepressant treatment response. Arch Gen Psychiatry 2010; 67:369–379Crossref, Google Scholar
22 Yoshida K, Takahashi H, Higuchi H, Kamata M, Ito K, Sato K, Naito S, Shimizu T, Itoh K, Inoue K, Suzuki T, Nemeroff CB: Prediction of antidepressant response to milnacipran by norepinephrine transporter gene polymorphisms. Am J Psychiatry 2004; 161:1575–1580Crossref, Google Scholar
23 Uher R, Huezo-Diaz P, Perroud N, Smith R, Rietschel M, Mors O, Hauser J, Maier W, Kozel D, Henigsberg N, Barreto M, Placentino A, Dernovsek MZ, Schulze TG, Kalember P, Zobel A, Czerski PM, Larsen ER, Souery D, Giovannini C, Gray JM, Lewis CM, Farmer A, Aitchison KJ, McGuffin P, Craig I: Genetic predictors of response to antidepressants in the GENDEP project. Pharmacogenomics J 2009; 9:225–233Crossref, Google Scholar
24 Laje G, Allen AS, Akula N, Manji H, John Rush A, McMahon FJ: Genome-wide association study of suicidal ideation emerging during citalopram treatment of depressed outpatients. Pharmacogenet Genomics 2009; 19:666–674Crossref, Google Scholar
25 Garriock HA, Kraft JB, Shyn SI, Peters EJ, Yokoyama JS, Jenkins GD, Reinalda MS, Slager SL, McGrath PJ, Hamilton SP: A genomewide association study of citalopram response in major depressive disorder. Biol Psychiatry 2010; 67:133–138Crossref, Google Scholar
26 Ueda HR, Chen W, Adachi A, Wakamatsu H, Hayashi S, Takasugi T, Nagano M, Nakahama K, Suzuki Y, Sugano S, Iino M, Shigeyoshi Y, Hashimoto S. R., Chen W, Adachi A, Wakamatsu H, Hayashi S, Takasugi T, Nagano M, Nakahama K, Suzuki Y, Sugano S, Iino M, Shigeyoshi Y, Hashimoto S: A transcription factor response element for gene expression during circadian night. Nature 2002; 418:534–539Crossref, Google Scholar
27 Ising M, Lucae S, Binder EB, Bettecken T, Uhr M, Ripke S, Kohli MA, Hennings JM, Horstmann S, Kloiber S, Menke A, Bondy B, Rupprecht R, Domschke K, Baune BT, Arolt V, Rush AJ, Holsboer F, Müller-Myhsok B: A genomewide association study points to multiple loci that predict antidepressant drug treatment outcome in depression. Arch Gen Psychiatry 2009; 66:966–975Crossref, Google Scholar
28 Uher R, Perroud N, Ng MY, Hauser J, Henigsberg N, Maier W, Mors O, Placentino A, Rietschel M, Souery D, Zagar T, Czerski PM, Jerman B, Larsen ER, Schulze TG, Zobel A, Cohen-Woods S, Pirlo K, Butler AW, Muglia P, Barnes MR, Lathrop M, Farmer A, Breen G, Aitchison KJ, Craig I, Lewis CM, McGuffin P: Genome-wide pharmacogenetics of antidepressant response in the GENDEP project. Am J Psychiatry 2010; 167:555–564Crossref, Google Scholar
29 Moldin SO: Sponsoring initiatives in the molecular genetics of mental disorders, in Genetics and Mental Disorders Report of the NIMH Genetics Workgroup: NIH Publication 98-4268. Bethesda, Md, National Institutes of Health, 1998 (note Table 4)Google Scholar
30 Meltzer HY: Role of serotonin in the action of atypical antipsychotic drugs. Clin Neurosci 1995; 3:64–75Google Scholar
31 Meltzer HY, Li Z, Kaneda Y, Ichikawa J: Serotonin receptors: their key role in drugs to treat schizophrenia. Prog Neuropsychopharmacol Biol Psychiatry 2003; 27:1159–1172Crossref, Google Scholar
32 Stöber G, Jatzke S, Heils A, Jungkunz G, Fuchs E, Knapp M, Riederer P, Lesch KP. Susceptibility for schizophrenia is not influenced by a functional insertion/deletion variant in the promoter of the serotonin transporter gene. Eur Arch Psychiatry Clin Neurosci 1998; 248:82–86Crossref, Google Scholar
33 Konneker TI, Crowley JJ, Quackenbush CR, Keefe RS, Perkins DO, Stroup TS, Lieberman JA, van den Oord E, Sullivan PF: No association of the serotonin transporter polymorphisms 5-HTTLPR and RS25531 with schizophrenia or neurocognition. Am J Med Genet B Neuropsychiatr Genet 2010; 153B:1115–1117Google Scholar
34 Lohmueller KE, Pearce CL, Pike M, Lander ES, Hirschhorn JN: Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nat Genet 2003; 33:177–182Crossref, Google Scholar
35 Spurlock G, Heils A, Holmans P, Williams J, D'Souza UM, Cardno A, Murphy KC, Jones L, Buckland PR, McGuffin P, Lesch KP, Owen MJ: A family based association study of T102C polymorphism in 5HT2A and schizophrenia plus identification of new polymorphisms in the promoter. Mol Psychiatry 1998; 3:42–49Crossref, Google Scholar
36 Kim B, Choi EY, Kim CY, Song K, Joo YH: Could HTR2A T102C and DRD3 Ser9Gly predict clinical improvement in patients with acutely exacerbated schizophrenia? Results from treatment responses to risperidone in a naturalistic setting. Hum Psychopharmacol 2008; 23:61–67Crossref, Google Scholar
37 Mössner R, Schuhmacher A, Kühn KU, Cvetanovska G, Rujescu D, Zill P, Quednow BB, Rietschel M, Wölwer W, Gaebel W, Wagner M, Maier W: Functional serotonin 1A receptor variant influences treatment response to atypical antipsychotics in schizophrenia. Pharmacogenet Genomics 2009; 19:91–94Crossref, Google Scholar
38 Wang L, Fang C, Zhang A, Du J, Yu L, Ma J, Feng G, Xing Q, He L: The −1019 C/G polymorphism of the 5-HT1A receptor gene is associated with negative symptom response to risperidone treatment in schizophrenia patients. J Psychopharmacol 2008; 22:904–909Crossref, Google Scholar
39 Reynolds GP, Yao Z, Zhang X, Sun J, Zhang Z: Pharmacogenetics of treatment in first-episode schizophrenia: D3 and 5-HT2C receptor polymorphisms separately associate with positive and negative symptom response. Eur Neuropsychopharmacol 2005; 15:143–151Crossref, Google Scholar
40 Souza RP, de Luca V, Meltzer HY, Lieberman JA, Kennedy JL: Influence of serotonin 3A and 3B receptor genes on clozapine treatment response in schizophrenia. Pharmacogenet Genomics 2010; 20:274–276Google Scholar
41 Murphy KC, Jones LA, Owen MJ: High rates of schizophrenia in adults with velo-cardio-facial syndrome. Arch Gen Psychiatry 1999; 56:940–945Crossref, Google Scholar
42 Egan MF, Goldberg TE, Kolachana BS, Callicott JH, Mazzanti CM, Straub RE, Goldman D, Weinberger DR: Effect of COMT Val108/158 Met genotype on frontal lobe function and risk for schizophrenia. Proc Natl Acad Sci USA 2001; 98:6917–6922Crossref, Google Scholar
43 Bray NJ, Buckland PR, Williams NM, Williams HJ, Norton N, Owen MJ, O'Donovan MC: A haplotype implicated in schizophrenia susceptibility is associated with reduced COMT expression in human brain. Am J Hum Genet 2003; 73:152–161Crossref, Google Scholar
44 Palmatier MA, Kang AM, Kidd KK: Global variation in the frequencies of functionally different catechol-O-methyltransferase alleles. Biol Psychiatry 1999; 46:557–567Crossref, Google Scholar
45 Lee SG, Joo Y, Kim B, Chung S, Kim HL, Lee I, Choi B, Kim C, Song K: Association of Ala72Ser polymorphism with COMT enzyme activity and the risk of schizophrenia in Koreans. Hum Genet 2005; 116:319–328Crossref, Google Scholar
46 Anttila S, Illi A, Kampman O, Mattila KM, Lehtimäki T, Leinonen E: Interaction between NOTCH4 and catechol-O-methyltransferase genotypes in schizophrenia patients with poor response to typical neuroleptics. Pharmacogenetics 2004; 14:303–307Crossref, Google Scholar
47 Gupta M, Bhatnagar P, Grover S, Kaur H, Baghel R, Bhasin Y, Chauhan C, Verma B, Manduva V, Mukherjee O, Purushottam M, Sharma A, Jain S, Brahmachari SK, Kukreti R: Association studies of catechol-O-methyltransferase (COMT) gene with schizophrenia and response to antipsychotic treatment. Pharmacogenomics 2009; 10:385–397Crossref, Google Scholar
48 Kang CY, Xu XF, Shi ZY, Yang JZ, Liu H, Xu HH: Interaction of catechol-O-methyltransferase (COMT) Val108/158 Met genotype and risperidone treatment in Chinese Han patients with schizophrenia. Psychiatry Res 2010; 176:94–95Crossref, Google Scholar
49 Olney JW, Newcomer JW, Farber NB: NMDA receptor hypofunction model of schizophrenia. J Psychiatr Res 1999; 33:523–533Crossref, Google Scholar
50 Millar JK, Wilson-Annan JC, Anderson S, Christie S, Taylor MS, Semple CA, Devon RS, St Clair DM, Muir WJ, Blackwood DH, Porteous DJ: Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum Mol Genet 2000; 9:1415–1423Crossref, Google Scholar
51 Hayashi-Takagi A, Takaki M, Graziane N, Seshadri S, Murdoch H, Dunlop AJ, Makino Y, Seshadri AJ, Ishizuka K, Srivastava DP, Xie Z, Baraban JM, Houslay MD, Tomoda T, Brandon NJ, Kamiya A, Yan Z, Penzes P, and Sawa A: Disrupted-in-Schizophrenia 1 (DISC1) regulates spines of the glutamate synapse via Rac1. Nat Neurosci 2010; 13:327–332Crossref, Google Scholar
52 Mouaffak F, Kebir O, Chayet M, Tordjman S, Vacheron MN, Millet B, Jaafari N, Bellon A, Olié JP, Krebs MO: Association of Disrupted in Schizophrenia 1 (DISC1) missense variants with ultra-resistant schizophrenia. Pharmacogenomics J 2010; doi:https://doi.org/10.1038/tpj.2010.40Google Scholar
53 McClay JL, Adkins DE, Aberg K, Stroup S, Perkins DO, Vladimirov VI, Lieberman JA, Sullivan PF, van den Oord EJ: Genome-wide pharmacogenomic analysis of response to treatment with antipsychotics. Mol Psychiatry 2009; doi:https://doi.org/10.1038/mp.2009.89Google Scholar
54 Lavedan C, Licamele L, Volpi S, Hamilton J, Heaton C, Mack K, Lannan R, Thompson A, Wolfgang CD, Polymeropoulos MH: Association of the NPAS3 gene and five other loci with response to the antipsychotic iloperidone identified in a whole genome association study. Mol Psychiatry 2009; 14:804–819Crossref, Google Scholar
55 Ikeda M, Tomita Y, Mouri A, Koga M, Okochi T, Yoshimura R, Yamanouchi Y, Kinoshita Y, Hashimoto R, Williams HJ, Takeda M, Nakamura J, Nabeshima T, Owen MJ, O'Donovan MC, Honda H, Arinami T, Ozaki N, Iwata N: Identification of novel candidate genes for treatment response to risperidone and susceptibility for schizophrenia: integrated analysis among pharmacogenomics, mouse expression, and genetic case-control association approaches. Biol Psychiatry 2010; 67:263–269Crossref, Google Scholar
56 Loerch PM, Lu T, Dakin KA, Vann JM, Isaacs A, Geula C, Wang J, Pan Y, Gabuzda DH, Li C, Prolla TA, Yankner BA: Evolution of the aging brain transcriptome and synaptic regulation. PLoS One 2008; 3:e3329Crossref, Google Scholar
57 Alkelai A, Greenbaum L, Rigbi A, Kanyas K, Lerer B: Genome-wide association study of antipsychotic-induced parkinsonism severity among schizophrenia patients. Psychopharmacology (Berl) 2009; 206:491–499Crossref, Google Scholar
58 Aberg K, Adkins DE, Bukszár J, Webb BT, Caroff SN, Miller del D, Sebat J, Stroup S, Fanous AH, Vladimirov VI, McClay JL, Lieberman JA, Sullivan PF, van den Oord EJ: Genomewide association study of movement-related adverse antipsychotic effects. Biol Psychiatry 2010; 67:279–282Crossref, Google Scholar
59 Inada T, Koga M, Ishiguro H, Horiuchi Y, Syu A, Yoshio T, Takahashi N, Ozaki N, Arinami T: Pathway-based association analysis of genome-wide screening data suggest that genes associated with the γ-aminobutyric acid receptor signaling pathway are involved in neuroleptic-induced, treatment-resistant tardive dyskinesia. Pharmacogenet Genomics 2008; 18:317–323Crossref, Google Scholar
60 Syu A, Ishiguro H, Inada T, Horiuchi Y, Tanaka S, Ishikawa M, Arai M, Itokawa M, Niizato K, Iritani S, Ozaki N, Takahashi M, Kakita A, Takahashi H, Nawa H, Keino-Masu K, Arikawa-Hirasawa E, Arinami T: Association of the HSPG2 gene with neuroleptic-induced tardive dyskinesia. Neuropsychopharmacology 2010; 35:1155–1164Crossref, Google Scholar
61 Kendler KS, Pedersen NL, Neale MC, Mathé AA: A pilot Swedish twin study of affective illness including hospital- and population-ascertained subsamples: results of model fitting. Behav Genet 1995; 25:217–232Crossref, Google Scholar
62 McGuffin P, Rijsdijk F, Andrew M, Sham P, Katz R, Cardno A: The heritability of bipolar affective disorder and the genetic relationship to unipolar depression. Arch Gen Psychiatry 2003; 60:497–502Crossref, Google Scholar
63 Kieseppä T, Partonen T, Haukka J, Kaprio J, Lönnqvist J: High concordance of bipolar I disorder in a nationwide sample of twins. Am J Psychiatry 2004; 161:1814–1821Crossref, Google Scholar
64 Perlis RH, Smoller JW, Ferreira MA, McQuillin A, Bass N, Lawrence J, Sachs GS, Nimgaonkar V, Scolnick EM, Gurling H, Sklar P, Purcell S. A genome-wide association study of response to lithium for prevention of recurrence in bipolar disorder. Am J Psychiatry 2009; 166:718–725Crossref, Google Scholar
65 Tucker RP, Chiquet-Ehrismann R: Teneurins: a conserved family of transmembrane proteins involved in intercellular signaling during development Dev Biol 2006; 290:237–245Crossref, Google Scholar
66 Ingelman-Sundberg M: Genetic polymorphisms of cytochrome P450 2D6 (CYP2D6): clinical consequences, evolutionary aspects and functional diversity. Pharmacogenomics J 2005; 5:6–13Crossref, Google Scholar
67 Nebert DW, Dieter MZ: The evolution of drug metabolism. Pharmacology 2000; 61:124–135Crossref, Google Scholar
68 de Leon J, Armstrong SC, Cozza KL: Clinical guidelines for psychiatrists for the use of pharmacogenetic testing for CYP450 2D6 and CYP450 2C19. Psychosomatics 2006; 47:75–85Crossref, Google Scholar
69 Brosen K: Some aspects of genetic polymorphism in the biotransformation of antidepressants. Therapiewoche 2004; 59:5–12Google Scholar
70 Arranz MJ, de Leon J: Pharmacogenetics and pharmacogenomics of schizophrenia: a review of last decade of research. Mol Psychiatry 2007; 12:707–747Crossref, Google Scholar
71 de Leon J: AmpliChip CYP450 test: personalized medicine has arrived in psychiatry. Expert Rev Mol Diagn 2006; 6:277–286Crossref, Google Scholar
72 Evaluation of Genomic Applications in Practice and Prevention (EGAPP) Working Group: Recommendations from the EGAPP Working Group: testing for cytochrome P450 polymorphisms in adults with nonpsychotic depression treated with selective serotonin reuptake inhibitors. Genet Med 2007; 9:819–825Crossref, Google Scholar
73 Thakur M, Grossman I, McCrory DC, Orlando LA, Steffens DC, Cline KE, Gray RN, Farmer J, DeJesus G, O'Brien C, Samsa G, Goldstein DB, Matchar DB: Review of evidence for genetic testing for CYP450 polymorphisms in management of patients with nonpsychotic depression with selective serotonin reuptake inhibitors. Genet Med 2007; 9:826–835Crossref, Google Scholar
74 Heller T, Kirchheiner J, Armstrong VW, Luthe H, Tzvetkov M, Brockmöller J, Oellerich M: AmpliChip CYP450 GeneChip: a new gene chip that allows rapid and accurate CYP2D6 genotyping. Ther Drug Monit 2006; 28:673–677Crossref, Google Scholar
75 Dunbar L, Butler R, Wheeler A, Pulford J, Miles W, Sheridan J: Clinician experiences of employing the AmpliChip\R CYP450 test in routine psychiatric practice. J Psychopharmacol 2009; doi: https://doi.org/10.1177/0269881109106957Google Scholar
76 Cai WM, Nikoloff DM, Pan RM, de Leon J, Fanti P, Fairchild M, Koch WH, and Wedlund PJ: CYP2D6 genetic variation in healthy adults and psychiatric African-American subjects: implications for clinical practice and genetic testing. Pharmacogenomics J 2006; 6:343–350Crossref, Google Scholar