Schizophrenia Susceptibility Genes: Emergence of Positional Candidates and Future Directions
Abstract
Schizophrenia is a devastating psychiatric disorder that affects ~1% of the population worldwide. It is characterized by so-called ‘positive symptoms’–including delusions and hallucinations–‘negative symptoms’–including blunted emotions and social isolation–and cognitive deficits–including impairments in attention and working memory. Studies of the inheritance of schizophrenia have revealed that it is a multifactorial disease that is characterized by multiple genetic susceptibility elements, each contributing a modest degree of risk. Linkage studies have identified several potential schizophrenia susceptibility loci, and in recent years major progress has been made in the identification of positional candidate susceptibility genes from these loci. A central goal of future research will be to use this genetic knowledge to generate specific animal models, characterize genetic interactions, investigate the disease pathophysiology and assist drug-discovery efforts.
The genetic component of schizophrenia
Schizophrenia is a severe psychiatric disorder that has a lifetime prevalence of ~1% in most of the populations studied (1). Similar to many common complex disorders, schizophrenia is a multifactorial disorder that is characterized by the contribution of multiple susceptibility genes that could act in conjunction with epigenetic processes and environmental factors (1). More than 20 genome-wide scans aiming to localize genes for this disorder have been reported. Two recent meta-analyses (2, 3) of the combined results of several genome-wide studies were performed to attempt to clarify inconsistencies among individual studies (meta-analytic approaches tend to amplify signals that are weak but consistent among studies and to attenuate signals that are strong but non-reproducible). One meta-analysis study confirmed 8p, 13q and 22q as valid linkage regions that probably contain one or more susceptibility genes. The second meta-analysis study (using a different statistical methodology) implicated 2p12–q22.1 (under stringent criteria), in addition to loci at 5q, 3p, 11q, 2q, 1q, 22q, 8p, 6p, 20p and 14q (under less-stringent thresholds). One interpretation of these findings is that approximately ten regions of the genome are likely to contain schizophrenia susceptibility genes, although this is almost certainly an underestimate because: (i) meta-analytic approaches tend to attenuate true strong signals that are population specific; and (ii) it is expected that many schizophrenia susceptibility genes are undetectable using linkage studies. The ultimate validation of the linkage results will be the identification of the susceptibility genes themselves. Indeed, in the past three years, the field of schizophrenia genetics has moved to the systematic positional cloning of susceptibility genes from chromosomal regions that were first identified by linkage approaches. These systematic efforts employ the genotyping of relatively large numbers of markers, including single nucleotide polymorphisms (SNPs) and linkage disequilibrium (LD) assays in family-based or case–control samples, and have resulted in the identification of strong positional candidate genes.
In this article, we discuss the genetic data regarding these strong positional candidate genes that were identified through the systematic follow-up of linkage signals (in chronological order of publication of the reports), in addition to their possible biological functions. We also discuss the genetic data for three candidate genes that are located in the vicinity of linkage signals and are identified through multipronged candidate gene approaches, rather than systematic positional cloning approaches. Finally, we briefly comment on the statistical support for these findings and the future directions of genetic research in the context of advancing the understanding of how genetic factors contribute biologically to the disease process.
Owing to space limitations, other genes that could be good candidates (e.g. DRD3, CHRNA2, BDNF, GAD2 and AKT1) but do not conform to the criteria outlined in the previous paragraph are not discussed in this article. Furthermore, we cite a limited number of follow-up genetic studies for each genetic finding, prioritizing the ones that use relatively large family-based samples. Such samples are considered more reliable than case–control samples in which factors such as hidden population stratification can confound the interpretation of a positive or negative finding (4).
Genes identified through the systematic follow-up of linkage signals
The first report of a strong positional candidate schizophrenia gene that was identified by a systematic fine-mapping approach within a region implicated by linkage analysis was published in 2002 (5). Soon thereafter, three additional reports described three new susceptibility genes that were identified using similar approaches (6–8). More recently, additional genes have been reported based on the systematic SNP-based follow-up analysis of linkage peaks (9–14) (Table 1).
Proline dehydrogenase
The proline dehydrogenase (PRODH) gene is located on chromosome 22q11, which is a region implicated by some linkage studies (2, 3) and also frequently deleted in patients with schizophrenia (15). Several studies have established that the risk of schizophrenia for a patient with a 22q11 microdeletion is ~25–31 times the general population risk of 1% (16, 17) and that the rate of 22q11 microdeletions in schizophrenia, although relatively low, is ~12–80 times the estimated general population rate (15). This first unequivocal association between a well-defined genetic lesion and schizophrenia facilitated fine-mapping efforts at this locus. LD analysis in family samples (triads) that tested for preferential transmission of 72 SNPs and multi-SNP haplotypes from parents to affected (non-deleted) individuals identified the over-transmission of a gene variant located at the 30 end of the PRODH gene (5, 18). This finding was recently replicated in two independent family-based samples, including a large collection of 528 families from China (19) and 274 families of Ashkenazi Jewish origin (20), although one negative family study has also been reported (21). Moreover, 30-end variants of the gene were also identified as being a risk factor for the development of psychotic symptoms during adolescence in children with 22q11 microdeletions (22). Although the implicated variants are consistently located at the 3′ end of the gene, their functional consequences are still unknown. However, rare variants of the PRODH gene affecting highly conserved amino acids (generated through gene conversion from a nearby pseudogene) that are enriched to various degrees in samples of individuals with schizophrenia have been identified (5). This discovery gained additional support in an independent set of studies (23, 24), and functional analysis has linked several of these variants with marked reductions in enzymatic activity (25). PRODH encodes an enzyme that metabolizes l-proline—a putative neuromodulatory amino acid that could directly influence glutamate-mediated transmission, which is believed to have a crucial role in the pathophysiology of schizophrenia (26). In addition, Prodh-deficient mice show dysregulation of cortical dopamine turnover and transmission that is reminiscent of schizophrenia in humans (26).
Dystrobrevin-binding protein 1
The dystrobrevin-binding protein 1, or dysbindin, (DTNBP1) gene maps within a broad region on chromosome 6p where there is evidence of linkage to schizophrenia in Irish families (27). Genetic variants of this gene are associated with schizophrenia (6) in the same families. This finding has been replicated in several additional samples, including some family-based samples, but negative studies have also been reported and considerable putative allelic heterogeneity was evident among the positive studies (28–32). Initial expression and functional studies provide some additional support for a role for DTNBP1 in schizophrenia. DTNBP1 is a member of the biogenesis of lysosome-related organelles complex (33) and the dystrophin protein complex (34). It has a widespread distribution in the brain, including expression in pyramidal neurons in the hippocampus and the dorsolateral pre-frontal cortex (DLPFC). Two recent studies provide evidence that DTNBP1 expression is decreased in schizophrenia in both the DLPFC and the excitatory pathways of the hippocampus (35, 36). A substantial fraction of DTNBP1 is presynaptically localized, and preliminary in vitro evidence suggests that knockdown of endogenous dysbindin protein results in the reduction of presynaptic protein expression and glutamate release, indicating that dysbindin might influence exocytotic glutamate release (37).
Neuregulin 1
Neuregulin 1 (NRG1) was identified as being a susceptibility gene for schizophrenia following a genome-wide linkage scan of 33 Icelandic families with schizophrenia that highlighted a locus on chromosome 8p (7). Fine-mapping of the 8p locus, together with haplotype association analysis of a large number of patients with schizophrenia and control individuals, narrowed the region of interest to the 5′ end of the NRG1 gene. A core haplotype at the 50 end of the gene comprising several markers within a 290-kb LD block showed highly significant association with schizophrenia (7). The functional consequences of this gene variant are still unknown. NRG1 association with schizophrenia has been observed in several additional samples, including some reliable family-based samples, although considerable allelic heterogeneity was evident in these studies (38–41). Negative studies have also been reported (20, 42, 43). The NRG1 gene encodes a well-characterized protein that is involved in many neuronal functions, ranging from neuronal survival to myelination and synaptic plasticity (44).
G72
The G72 gene is located within a broad linkage peak that extends from 13q32 to q34, where there is evidence of linkage to both schizophrenia and bipolar disorder (2, 45). Significant association with schizophrenia was observed for several SNPs and haplotypes at the G72 locus in a French–Canadian case–control sample, and the association for two SNPs was replicated in a Russian case–control cohort (8). Interestingly, a subsequent study provided evidence of an association between variants at the G72 locus and bipolar disorder (46). The association of G72 with schizophrenia has been observed in several additional samples with evidence of allelic heterogeneity, although negative studies have also been reported (47–53). Expression and functional studies indicate a potential interaction between G72 and D-amino acid oxidase (DAAO) that modulates the DAAO enzymatic activity and, thus, could indirectly affect glutamate-mediated signaling (8, 54). However, this interaction remains to be demonstrated in vivo.
Disrupted in schizophrenia 1
A balanced translocation involving chromosomes 1 and 11 (1q42.1;11q14.3) was strongly linked to psychopathology, including schizophrenia, depression and mania in a large Scottish family. The 1q breakpoint was cloned and was found to involve two genes: disrupted in schizophrenia (DISC)1 and DISC2 (the latter is a noncoding, presumably regulatory, RNA) (55). Although DISC1 was identified five years ago, it was a more recent large-scale linkage (56) and follow-up systematic LD analysis in families from Finland that identified DISC1 as being a positional candidate from the 1q42 locus (9). DISC1 association with schizophrenia has been observed in several additional samples with evidence of allelic heterogeneity, although negative studies have also been reported (20, 57, 58). Interestingly, a family afflicted with schizophrenia and schizoaffective disorder was recently shown to have segregated a rare frameshift variant of the gene (59). DISC1 is a complex gene, the involvement of which in development and synaptic plasticity is poorly understood. The protein it encodes is associated with numerous cytoskeletal proteins and could be involved in centrosomal and microtubule function, cell migration, neurite outgrowth, membrane trafficking of receptors, mitochondrial function and, possibly, phosphodiesterase function (60, 61).
C-terminal PDZ ligand of neuronal nitric oxide synthase
LD analysis, using 14 microsatellite markers and 15 SNPs from a subregion of a previously reported linkage locus at 1q22 (62), in large Canadian families with schizophrenia produced nominally significant evidence of LD between schizophrenia and a subset of markers that is located within the genomic extent of the C-terminal PDZ ligand of neuronal nitric oxide synthase (CAPON) gene, making CAPON a prime positional candidate from the schizophrenia susceptibility locus on 1q22 (10). An abnormal expression pattern of this gene was observed in the brains of individuals with schizophrenia or bipolar disorder (63). Two replication studies, one positive and one negative, have been reported (64, 65). CAPON is involved in NMDA-receptor-coupled nitric oxide signaling (66).
ZDHHC8
The involvement of this gene in schizophrenia was identified in the same LD screen of the 22q11 locus that led to the discovery of the PRODH–schizophrenia association (5, 18). More recently, it was shown that one of the ZDHHC8 risk alleles (at SNP rs175174), located in intron 4, affects the ratio of an intron-4-containing unspliced form (encoding a putative truncated inactive form of the protein) to the fully spliced active form (11). The relatively subtle consequent change in the level of the active protein led to ~1.5-fold increase in disease risk in two tested family samples (11). Other variants of the gene (affecting distinct aspects of its complex splicing or its expression level) could modulate the disease risk in other samples. One positive family-based study and one negative family-based study have been reported (67, 68). The effect of the gene is predicted to be much stronger in individuals with 22q11 deletions and schizophrenia, in which a 50% (or ~65% when the non-deleted allele carries, for example, the risk SNP rs175174 variant) decrease in ZDHHC8 activity levels is predicted. ZDHHC8 encodes a transmembrane palmitoyl-transferase that modifies, among other targets, postsynaptic density protein of 95 kDa (PSD-95) and could have an important role in excitatory synaptic transmission (69).
Trace amine receptor 4
Trace amine receptor 4 (TAAR6) was identified in an LD study (12) of European-ancestry and African–American families with schizophrenia that previously showed evidence of linkage to 6q13–q26 (70). This LD study (12) focused on subregion q23.2, which contains several functional candidate genes for schizophrenia. A primary screen using 31 SNPs and a follow-up higher-density screen using 23 SNPs over a 21.6-kb region highlighted TAAR6 (12) as being a prime positional candidate gene from the schizophrenia susceptibility locus on 6q23.2. Two negative replication studies have been reported (71, 72). However, an independent study (73) implicated TAAR6 (which is a G-protein-coupled receptor that is widely expressed in the brain (12, 74)) in susceptibility to bipolar disorder.
Epsin 4
Chromosome 5q33 is a region that has shown evidence of linkage to schizophrenia in four independent linkage studies. Four adjacent markers (and associated haplotypes) at the 5′ end of the epsin 4 (EPN4) gene, which is located in this region, showed significant evidence of LD with schizophrenia in a fine-mapping study that used 450 unrelated English, Irish, Welsh and Scottish research subjects with schizophrenia and 450 ancestrally matched supernormal controls (13). The EPN4 gene encodes the clathrin-associated protein enthoprotin, which has a role in the transport and stability of neurotransmitter vesicles at the synapses and within neurons (75). No replication studies have been reported.
γ-Aminobutyric acid A receptor subunit gene cluster
An early genome-wide linkage scan in Portuguese families with schizophrenia identified a risk locus on chromosome 5q31–q35 (76) – a finding supported by subsequent meta-analysis. A two-stage candidate gene association approach focused on a group of γ-aminobutyric acid (GABA)A receptor subunit genes (GABRA1, GABRA6, GABRB2, GABRG2 and GABRP) within this linkage peak (14). In the first stage, associations were detected in a Portuguese patient sample with SNPs and haplotypes in GABRA1, GABRP and GABRA6. The GABRA1 and GABRP findings were replicated in the second stage in an independent German family-based sample (14). These genes are plausible candidates based on prior speculation about the involvement of the GABA system in schizophrenia (77, 78).
Candidate genes located in the vicinity of linkage signals identified through candidate gene approaches
The candidacy of the genes described in this section is based on convergent genetic and biological evidence. Although unproven, the recurrent observation of the clustering of candidate susceptibility genes might indicate that more than one gene could contribute to at least some of the linkage signals in psychiatric disorders.
Catechol-O-methyltransferase
The catechol-O-methyltransferase (COMT) gene is located in the 22q11 locus between the PRODH and ZDHHC8 genes, and is a strong positional and functional candidate. COMT metabolizes released dopamine, and variation in COMT activity could have effects that are specific to the prefrontal cortex (PFC). This regionally selective effect of COMT might depend on the relatively low abundance and non-synaptic localization of the dopamine transporter in the PFC compared with the striatum (79). A high-activity form of the enzyme (Val158) was proposed to increase susceptibility to schizophrenia (79). Association studies using the clinical diagnosis of schizophrenia as phenotype are equivocal (80–86), although this form of the gene modulated executive function in some studies, which is affected in individuals with schizophrenia (79, 87). More-recent studies in animal models, however, indicate that low activity of this enzyme could be a risk factor for schizophrenia and that COMT might function as part of the genome buffering capacity to counteract the effect of other primary mutations that affect dopamine turnover and signaling in the frontal cortex (26). This prediction is supported by the results of a longitudinal follow-up study of children with 22q11 microdeletions. This study showed that the low-activity form of the enzyme (Met158) is a risk factor for decline in prefrontal cortical volume and cognition, and the consequent development of psychotic symptoms, during adolescence in these children (22). Overall, the potential contribution of COMT to schizophrenia is likely to be complex. In addition, the gene seems to have a functionally complex allelic architecture, with some alleles (Val158Met) determining the stability of the protein (88) and others determining the level of expression (86, 89).
Regulator of G-protein signaling 4
Regulator of G-protein signaling (RGS)4 was initially identified as being the only transcript (out of 7800 sampled by Mirnics et al. (90)) that was consistently reduced in the DLPFC of individuals with schizophrenia. The gene maps to 1q21–22, 0.7 Mb from CAPON. Chowdari et al. (91) genotyped 13 SNPs across a 300-kb segment spanning the gene in several independent datasets and found weak evidence of association with schizophrenia in each of the samples, although not in an allele-consistent manner. In most cases, association was present for a haplotype block stretching from intron 1 to several kilobase pairs upstream of the transcription start site. Independent replications have been reported but negative studies also exist (20, 92–94). Of the 19 human RGS transcripts, RGS4 shows the highest expression in the brain compared with all other tissues and is abundant in the cerebral cortex (95). RGS4 is a GTPase activator that desensitizes Gi/o and Gq and, thereby, negatively modulates G-protein-mediated signaling by dopamine, metabotropic glutamate and muscarinic receptors (96).
Calcineurin γ catalytic subunit
Forebrain-specific calcineurin-knockout mice were reported to have a spectrum of behavioral abnormalities related to altered behaviors observed in schizophrenia patients (97). Follow-up studies identified calcineurin γ catalytic subunit (PPP3CC) as a potential schizophrenia susceptibility gene (98) and led to the proposal that alterations in calcineurin signaling contribute to schizophrenia pathogenesis. In support of this proposal, the gene is downregulated in the hippocampus of individuals with schizophrenia (99). The genetic association was not replicated, however, in a sample of Ashkenazi Jewish nuclear families (20). PPP3CC is located at 8p21.3, 10 Mb from NRG1 but adjacent to previously described linkage signals (98). Calcineurin is a multifunctional calcium-dependent serine/threonine phosphatase that is centrally involved in many aspects of synaptic plasticity. It has particular roles in glutamate and dopamine signaling and their interactions, including the regulation of DARPP32, a molecular node of convergence between dopamine receptor 1 and NMDA receptor signaling pathways (97).
Statistical support and generalization of genetic findings
The statistical burden of proof is lower for genes identified through systematic follow-up of linkage signals compared with genes picked in an essentially random fashion, irrespective of their location relative to linkage signals (for details, see Ref. (100)). Nevertheless, support for at least some of the findings described (e.g. PRODH, DTNBP1, NRG1, G72, DISC1 and COMT) seems to be strong, based on a combination of criteria: the degree of statistical significance, the reproducibility of the associations in independent family samples, the identification of independent rare risk alleles and the consistent findings from animal model studies and endophenotype-based studies in humans.
However, it is too early to draw firm conclusions about the generalization of these findings among different samples and populations based on the published replication studies, primarily because of issues regarding the extent of coverage of the implicated loci, the structure of the samples used in replication studies and publication bias. One important issue of concern regards the structure of the tested replication samples. It is becoming increasingly clear that, when allele frequencies differ notably among subpopulations that are not represented equally among cases and controls (population stratification), unreliable results can be obtained (4). The possibility, therefore, that replication studies using case–control samples represent false positives (or negatives) must be considered seriously. This is a problem that is relevant to all common complex disorders but it is likely to be more pronounced in genetic studies of psychiatric disorders, which are confounded by a larger degree of phenotypic heterogeneity. In addition, several of the employed ‘replication’ samples have been used repeatedly in genetic association studies, making the issue of multiple-testing corrections extremely relevant. These are important considerations given some striking inconsistencies among the variant alleles and haplotypes implicated in replication studies for at least some of the genes. It should be noted, however, that such inconsistencies are sometimes observed even in more-reliable family-based samples and can be explained in some instances by the presence of distinct variations that affect different functional elements within the gene that have emerged independently on a more recent ancestral background.
Future directions of genetic research
Clearly, there are still several ‘orphan’ linkage loci that await the identification of positional candidate genes, a task that will be facilitated by the sequencing of the human genome. It is likely that additional genes will also be identified through the genome-wide association studies that are starting to be implemented. As more genes are identified, a goal of future research will be to understand the functional implications and interactions of the susceptibility genes and their variants in the context of schizophrenia. Genetic studies of endophenotypes (79) (provided they are designed to avoid all of the pitfalls described earlier that are associated with genetic studies of the clinical syndrome), in addition to biological data from animal model studies, promise to advance the understanding of the disease pathophysiology in the coming years.
The question of true biological interaction among susceptibility genes is also extremely important in the field of complex psychiatric genetics, and might ultimately be better answered primarily by using a combination of molecular-based and animal-model-based approaches, as suggested by recent studies (26,60). Understanding the interactions between individual susceptibility elements could eventually aid the specificity of diagnosis and lead to the design of custom therapies with fewer side-effects and more-positive long-term disease outcomes for patients with specific genetic predispositions (Figure 1).
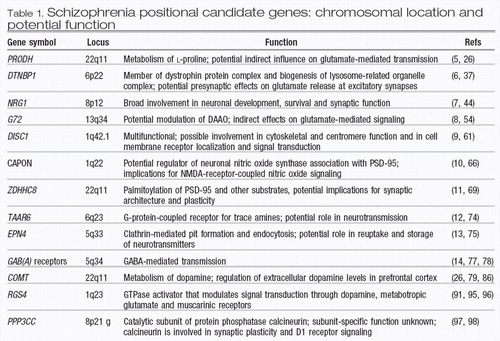 |
Table 1. Schizophrenia positional candidate genes: chromosomal location and potential function
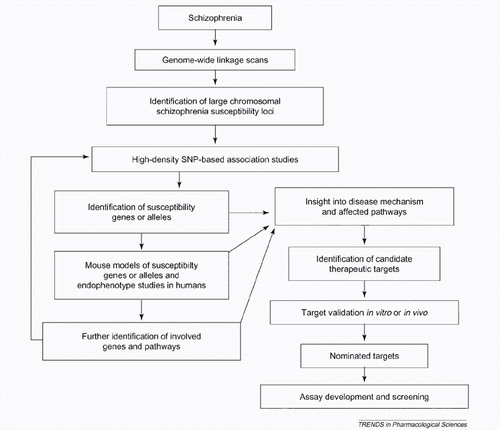
Figure 1. Flow diagram of the schizophrenia genetics research process and its potential application to drug discovery. In this scheme, the identification of susceptibility genes by SNP-based association studies, coupled with the generation and characterization of relevant mouse models and endophenotype studies in humans, forms a comprehensive system with which to identify the genes and molecular pathways involved in schizophrenia pathogenesis. This knowledge base provides a framework for mechanism-based drug-discovery efforts.
1 Karayiorgou, M. and Gogos, J.A. (1997) A turning point in schizophrenia genetics. Neuron 19, 967–979Crossref, Google Scholar
2 Badner, J.A. and Gershon, E.S. (2002) Meta-analysis of whole-genome linkage scans of bipolar disorder and schizophrenia. Mol. Psychiatry 7, 405–411Crossref, Google Scholar
3 Lewis, C.M. et al. (2003) Genome scan meta-analysis of schizophrenia and bipolar disorder, part II: schizophrenia. Am. J. Hum. Genet. 73, 34–48Crossref, Google Scholar
4 Campbell, C.D. et al. (2005) Demonstrating stratification in a European American population. Nat. Genet. 37, 868–872Crossref, Google Scholar
5 Liu, H. et al. (2002) Genetic variation at the 22q11 PRODH2/DGCR6 locus presents an unusual pattern and increases susceptibility to schizophrenia. Proc. Natl. Acad. Sci. U. S. A. 99, 3717–3722Crossref, Google Scholar
6 Straub, R.E. et al. (2002) Genetic variation in the 6p22.3 gene DTNBP1, the human ortholog of the mouse dysbindin gene, is associated with schizophrenia. Am. J. Hum. Genet. 71, 337–348Crossref, Google Scholar
7 Stefansson, H. et al. (2002) Neuregulin 1 and susceptibility to schizophrenia. Am. J. Hum. Genet. 71, 877–892Crossref, Google Scholar
8 Chumakov, I. et al. (2002) Genetic and physiological data implicating the new human gene G72 and the gene for d-amino acid oxidase in schizophrenia. Proc. Natl. Acad. Sci. U. S. A. 99, 13675–13680Crossref, Google Scholar
9 Hennah, W. et al. (2003) Haplotype transmission analysis provides evidence of association for DISC1 to schizophrenia and suggests sex-dependent effects. Hum. Mol. Genet. 12, 3151–3159Crossref, Google Scholar
10 Brzustowicz, L.M. et al. (2004) Linkage disequilibrium mapping of schizophrenia susceptibility to the CAPON region of chromosome 1q22. Am. J. Hum. Genet. 74, 1057–1063Crossref, Google Scholar
11 Mukai, J. et al. (2004) Evidence that the gene encoding ZDHHC8 contributes to the risk of schizophrenia. Nat. Genet. 36, 725–731Crossref, Google Scholar
12 Duan, J. et al. (2004) Polymorphisms in the trace amine receptor 4 (TRAR4) gene on chromosome 6q23.2 are associated with susceptibility to schizophrenia. Am. J. Hum. Genet. 75, 624–638Crossref, Google Scholar
13 Pimm, J. et al. (2005) The Epsin 4 gene on chromosome 5q, which encodes the clathrin-associated protein enthoprotin, is involved in the genetic susceptibility to schizophrenia. Am. J. Hum. Genet. 76, 902–907Crossref, Google Scholar
14 Petryshen, T.L. et al. (2005) Genetic investigation of chromosome 5q GABA(A) receptor subunit genes in schizophrenia. Mol. Psychiatry 10, 1074–1088Crossref, Google Scholar
15 Karayiorgou, M. et al. (1995) Schizophrenia susceptibility associated with interstitial deletions of chromosome 22q11. Proc. Natl. Acad. Sci. U. S. A. 92, 7612–7616Crossref, Google Scholar
16 Pulver, A.E. et al. (1994) Psychotic illness in patients diagnosed with velo-cardio-facial syndrome and their relatives. J. Nerv. Ment. Dis. 182, 476–478Crossref, Google Scholar
17 Murphy, K.C. et al. (1999) High rates of schizophrenia in adults with velo-cardio-facial syndrome. Arch. Gen. Psychiatry 56, 940–945Crossref, Google Scholar
18 Liu, H. et al. (2002) Genetic variation in the 22q11 locus and susceptibility to schizophrenia. Proc. Natl. Acad. Sci. U. S. A. 99, 16859–16864Crossref, Google Scholar
19 Li, T. et al. (2004) Evidence for association between novel polymorphisms in the PRODH gene and schizophrenia in a Chinese population. Am. J. Med. Genet. B Neuropsychiatr. Genet. 129B, 13–15Crossref, Google Scholar
20 Fallin, M.D. et al. (2005) Bipolar I disorder and schizophrenia: a 440-single-nucleotide polymorphism screen of 64 candidate genes among Ashkenazi Jewish case–parent trios. Am. J. Hum. Genet. 77, 918–936Crossref, Google Scholar
21 Williams, H.J. et al. (2003) Association between PRODH and schizophrenia is not confirmed. Mol. Psychiatry 8, 644–645Crossref, Google Scholar
22 Gothelf, D. et al. (2005) COMT genotype predicts longitudinal cognitive decline and psychosis in 22q11.2 deletion syndrome. Nat. Neurosci. 8, 1500–1502Crossref, Google Scholar
23 Jacquet, H. et al. (2002) PRODH mutations and hyperprolinemia in a subset of schizophrenic patients. Hum. Mol. Genet. 11, 2243–2249Crossref, Google Scholar
24 Jacquet, H. et al. (2005) Hyperprolinemia is a risk factor for schizoaffective disorder. Mol. Psychiatry 10, 479–485Crossref, Google Scholar
25 Bender, H.U. et al. (2005) Functional consequences of PRODH missense mutations. Am. J. Hum. Genet. 76, 409–420Crossref, Google Scholar
26 Paterlini, M. et al. (2005) Transcriptional and behavioral interaction between 22q11.2 orthologs modulates schizophrenia-related phenotypes in mice. Nat. Neurosci. 8, 1586–1594Crossref, Google Scholar
27 Straub, R.E. et al. (1995) A potential vulnerability locus for schizophrenia on chromosome 6p24–22: evidence for genetic heterogeneity. Nat. Genet. 11, 287–293Crossref, Google Scholar
28 Schwab, S.G. et al. (2003) Support for association of schizophrenia with genetic variation in the 6p22.3 gene, dysbindin, in sib-pair families with linkage and in an additional sample of triad families. Am. J. Hum. Genet. 72, 185–190Crossref, Google Scholar
29 Tang, J.X. et al. (2003) Family-based association study of DTNBP1 in 6p22.3 and schizophrenia. Mol. Psychiatry 8, 717–718Crossref, Google Scholar
30 Kirov, G. et al. (2004) Strong evidence for association between the dystrobrevin binding protein 1 gene (DTNBP1) and schizophrenia in 488 parent–offspring trios from Bulgaria. Biol. Psychiatry 55, 971–973Crossref, Google Scholar
31 Hall, D. et al. (2004) The contribution of three strong candidate schizophrenia susceptibility genes in demographically distinct populations. Genes Brain Behav. 3, 240–248Crossref, Google Scholar
32 De Luca, V. (2005) Untranslated region haplotype in dysbindin gene: analysis in schizophrenia. J. Neural Transm. 112, 1263–1267Crossref, Google Scholar
33 Li, W. et al. (2003) Hermansky-Pudlak syndrome type 7 (HPS-7) results from mutant dysbindin, a member of the biogenesis of lysosome-related organelles complex 1 (BLOC-1). Nat. Genet. 35, 84–89Crossref, Google Scholar
34 Benson, M.A. et al. (2001) Dysbindin, a novel coiled-coil-containing protein that interacts with the dystrobrevins in muscle and brain. J. Biol. Chem. 276, 24232–24241Crossref, Google Scholar
35 Talbot, K. et al. (2004) Dysbindin-1 is reduced in intrinsic, glutamatergic terminals of the hippocampal formation in schizophrenia. J. Clin. Invest. 113, 1353–1363Crossref, Google Scholar
36 Weickert, C.S. et al. (2004) Human dysbindin (DTNBP1) gene expression in normal brain and in schizophrenic prefrontal cortex. Arch. Gen. Psychiatry 61, 544–555Crossref, Google Scholar
37 Numakawa, T. et al. (2004) Evidence of novel neuronal functions of dysbindin, a susceptibility gene for schizophrenia. Hum. Mol. Genet. 13, 2699–2708Crossref, Google Scholar
38 Stefansson, H. et al. (2003) Association of neuregulin 1 with schizophrenia confirmed in a Scottish population. Am. J. Hum. Genet. 72, 83–87Crossref, Google Scholar
39 Yang, J.Z. et al. (2003) Association study of neuregulin 1 gene with schizophrenia. Mol. Psychiatry 8, 706–709Crossref, Google Scholar
40 Zhao, X. et al. (2004) A case control and family based association study of the neuregulin 1 gene and schizophrenia. J. Med. Genet. 41, 31–34Crossref, Google Scholar
41 Li, T. et al. (2004) Identification of a novel neuregulin 1 at-risk haplotype in Han schizophrenia Chinese patients, but no association with the Icelandic/Scottish risk haplotype. Mol. Psychiatry 9, 698–704Crossref, Google Scholar
42 Thiselton, D.L. et al. (2004) No evidence for linkage or association of neuregulin-1 (NRG1) with disease in the Irish study of high-density schizophrenia families (ISHDSF). Mol. Psychiatry 9, 777–783Crossref, Google Scholar
43 Duan, J. et al. (2005) Neuregulin 1 (NRG1) and schizophrenia: analysis of a US family sample and the evidence in the balance. Psychol. Med. 35, 1599–1610Crossref, Google Scholar
44 Corfas, G. et al. (2004) Neuregulin 1–erbB signaling and the molecular/ cellular basis of schizophrenia. Nat. Neurosci. 7, 575–580Crossref, Google Scholar
45 Detera-Wadleigh, S.D. et al. (1999) A high-density genome scan detects evidence for a bipolar-disorder susceptibility locus on 13q32 and other potential loci on 1q32 and 18p11.2. Proc. Natl. Acad. Sci. U. S. A. 96, 5604–5609Crossref, Google Scholar
46 Hattori, E. et al. (2003) Polymorphisms at the G72/G30 gene locus, on 13q33, are associated with bipolar disorder in two independent pedigree series. Am. J. Hum. Genet. 72, 1131–1140Crossref, Google Scholar
47 Schumacher, J. et al. (2004) Examination of G72 and D-amino-acid oxidase as genetic risk factors for schizophrenia and bipolar affective disorder. Mol. Psychiatry 9, 203–207Crossref, Google Scholar
48 Wang, X. et al. (2004) Association of G72/G30 with schizophrenia in the Chinese population. Biochem. Biophys. Res. Commun. 319, 1281–1286Crossref, Google Scholar
49 Addington, A.M. et al. (2004) Polymorphisms in the 13q32 gene G72/ G30 are associated with childhood-onset schizophrenia and psychosis not otherwise specified. Biol. Psychiatry 55, 976–980Crossref, Google Scholar
50 Korostishevsky, M. et al. (2004) Is the G72/G30 locus associated with schizophrenia? Single nucleotide polymorphisms, haplotypes, and gene expression analysis. Biol. Psychiatry 56, 169–176Crossref, Google Scholar
51 Korostishevsky, M. et al. (2006) Transmission disequilibrium and haplotype analyses of the G72/G30 locus: suggestive linkage to schizophrenia in Palestinian Arabs living in the north of Israel. Am. J. Med. Genet. B Neuropsychiatr. Genet. 141, 91–95Crossref, Google Scholar
52 Mulle, J.G. et al. (2005) No evidence for association to the G72/G30 locus in an independent sample of schizophrenia families. Mol. Psychiatry 10, 431–433Crossref, Google Scholar
53 Zou, F. et al. (2005) A family-based study of the association between the G72/G30 genes and schizophrenia in the Chinese population. Schizophr. Res. 73, 257–261Crossref, Google Scholar
54 Mothet, J.P. et al. (2000) d-serine is an endogenous ligand for the glycine site of the N-methyl-d-aspartate receptor. Proc. Natl. Acad. Sci. U. S. A. 97, 4926–4931Crossref, Google Scholar
55 Millar, J.K. et al. (2000) Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum. Mol. Genet. 9, 1415–1423Crossref, Google Scholar
56 Ekelund, J. et al. (2001) Chromosome 1 loci in Finnish schizophrenia families. Hum. Mol. Genet. 10, 1611–1617Crossref, Google Scholar
57 Callicott, J.H. et al. (2005) Variation in DISC1 affects hippocampal structure and function and increases risk for schizophrenia. Proc. Natl. Acad. Sci. U. S. A. 102, 8627–8632Crossref, Google Scholar
58 Hodgkinson, C.A. et al. (2004) Disrupted in schizophrenia 1 (DISC1): association with schizophrenia, schizoaffective disorder, and bipolar disorder. Am. J. Hum. Genet. 75, 862–872Crossref, Google Scholar
59 Sachs, N.A. et al. (2005) A frameshift mutation in disrupted in schizophrenia 1 in an American family with schizophrenia and schizoaffective disorder. Mol. Psychiatry 10, 758–764Crossref, Google Scholar
60 Millar, J.K. et al. (2005) DISC1 and PDE4B are interacting genetic factors in schizophrenia that regulate cAMP signaling. Science 310, 1187–1191Crossref, Google Scholar
61 Kamiya, A. et al. (2005) A schizophrenia-associated mutation of DISC1 perturbs cerebral cortex development. Nat. Cell Biol. 7, 1067–1078Crossref, Google Scholar
62 Brzustowicz, L.M. et al. (2000) Location of a major susceptibility locus for familial schizophrenia on chromosome 1q21–q22. Science 288, 678–682Crossref, Google Scholar
63 Xu, B. et al. (2005) Increased expression in dorsolateral prefrontal cortex of CAPON in schizophrenia and bipolar disorder. PLoSMed. 2, e263Google Scholar
64 Puri, V. et al. (2006) Failure to confirm allelic association between markers at the CAPON gene locus and schizophrenia in a British sample. Biol. Psychiatry 59, 195–197Crossref, Google Scholar
65 Zheng, Y. et al. (2005) Association of the carboxyl-terminal PDZ ligand of neuronal nitric oxide synthase gene with schizophrenia in the Chinese Han population. Biochem. Biophys. Res. Commun. 328, 809–815Crossref, Google Scholar
66 Jaffrey, S.R. et al. (1998) CAPON: a protein associated with neuronal nitric oxide synthase that regulates its interactions with PSD95. Neuron 20, 115–124Crossref, Google Scholar
67 Chen, W.Y. et al. (2004) Case–control study and transmission disequilibrium test provide consistent evidence for association between schizophrenia and genetic variation in the 22q11 gene ZDHHC8. Hum. Mol. Genet. 13, 2991–2995Crossref, Google Scholar
68 Glaser, B. et al. (2005) No association between the putative functional ZDHHC8 single nucleotide polymorphism rs175174 and schizophrenia in large European samples. Biol. Psychiatry 58, 78–80Crossref, Google Scholar
69 el-Husseini, A-D. and Bredt, D.S. (2002) Protein palmitoylation: a regulator of neuronal development and function. Nat. Rev. Neurosci. 3, 791–802Crossref, Google Scholar
70 Levinson, D.F. et al. (2000) Multicenter linkage study of schizophrenia candidate regions on chromosomes 5q, 6q, 10p, and 13q: schizophrenia linkage collaborative group III. Am. J. Hum. Genet. 67, 652–663Crossref, Google Scholar
71 Duan, S. et al. (2006) Failure to find association between TRAR4 and schizophrenia in the Chinese Han population. J. Neural Transm. 113, 381–385Crossref, Google Scholar
72 Ikeda, M. et al. (2005) No association of haplotype-tagging SNPs in TRAR4 with schizophrenia in Japanese patients. Schizophr. Res. 78, 127–130Crossref, Google Scholar
73 Abou Jamra, R. et al. (2005) A family-based and case–control association study of trace amine receptor genes on chromosome 6q23 in bipolar affective disorder. Mol. Psychiatry 10, 618–620Crossref, Google Scholar
74 Borowsky, B. et al. (2001) Trace amines: identification of a family of mammalian G protein-coupled receptors. Proc. Natl. Acad. Sci. U. S. A. 98, 8966–8971Crossref, Google Scholar
75 Wasiak, S. et al. (2002) Enthoprotin: a novel clathrin-associated protein identified through subcellular proteomics. J. Cell Biol. 158, 855–862Crossref, Google Scholar
76 Sklar, P. et al. (2004) Genome-wide scan in Portuguese Island families identifies 5q31–5q35 as a susceptibility locus for schizophrenia and psychosis. Mol. Psychiatry 9, 213–218Crossref, Google Scholar
77 Roberts, E. (1972) Prospects for research on schizophrenia. A hypothesis suggesting that there is a defect in the GABA system in schizophrenia. Neurosci. Res. Program Bull. 10, 468–482Google Scholar
78 Lewis, D.A. et al. (2005) Cortical inhibitory neurons and schizophrenia. Nat. Rev. Neurosci. 6, 312–324Crossref, Google Scholar
79 Egan, M.F. et al. (2001) Effect of COMT Val108/158 Met genotype on frontal lobe function and risk for schizophrenia. Proc. Natl. Acad. Sci. U. S. A. 98, 6917–6922Crossref, Google Scholar
80 Lohmueller, K.E. et al. (2003) Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nat. Genet. 33, 177–182Crossref, Google Scholar
81 Fan, J.B. et al. (2005) Catechol-O-methyltransferase gene Val/Met functional polymorphism and risk of schizophrenia: a large-scale association study plus meta-analysis. Biol. Psychiatry 57, 139–144Crossref, Google Scholar
82 Glatt, S.J. et al. (2003) Association between a functional catechol O-methyltransferase gene polymorphism and schizophrenia: meta-analysis of case–control and family-based studies. Am. J. Psychiatry 160, 469–476Crossref, Google Scholar
83 Munafo, M.R. et al. (2005) Lack of association of the COMT (Val158/108 Met) gene and schizophrenia: a meta-analysis of case–control studies. Mol. Psychiatry 10, 765–770Crossref, Google Scholar
84 Williams, H.J. et al. (2005) No association between schizophrenia and polymorphisms in COMT in two large samples. Am. J. Psychiatry 162, 1736–1738Crossref, Google Scholar
85 Tsai, S.J. et al. (2006) Lack of association of catechol-O-methyltransferase gene Val108/158Met polymorphism with schizophrenia: a family-based association study in a Chinese population. Mol. Psychiatry 11, 2–3Crossref, Google Scholar
86 Shifman, S. et al. (2002) A highly significant association between a COMT haplotype and schizophrenia. Am. J. Hum. Genet. 71, 1296–1302Crossref, Google Scholar
87 Ho, B.C. et al. (2005) Catechol-O-methyl transferase Val(158)Met gene polymorphism in schizophrenia: working memory, frontal lobe MRI morphology and frontal cerebral blood flow. Mol. Psychiatry 10, 287–298Crossref, Google Scholar
88 Chen, J. et al. (2004) Functional analysis of genetic variation in catechol-O-methyltransferase (COMT): effects on mRNA, protein, and enzyme activity in postmortem human brain. Am. J. Hum. Genet. 75, 807–821Crossref, Google Scholar
89 Bray, N.J. et al. (2003) A haplotype implicated in schizophrenia susceptibility is associated with reduced COMT expression in human brain. Am. J. Hum. Genet. 73, 152–161Crossref, Google Scholar
90 Mirnics, K. et al. (2000) Molecular characterization of schizophrenia viewed by microarray analysis of gene expression in prefrontal cortex. Neuron 28, 53–67Crossref, Google Scholar
91 Chowdari, K.V. et al. (2002) Association and linkage analyses of RGS4 polymorphisms in schizophrenia. Hum. Mol. Genet. 11, 1373–1380Crossref, Google Scholar
92 Chen, X. et al. (2004) Regulator of G-protein signaling 4 (RGS4) gene is associated with schizophrenia in Irish high density families. Am. J. Med. Genet. B Neuropsychiatr. Genet. 129, 23–26Crossref, Google Scholar
93 Zhang, F. et al. (2005) Association analysis of the RGS4 gene in Han Chinese and Scottish populations with schizophrenia. Genes Brain Behav. 4, 444–448Crossref, Google Scholar
94 Sobell, J.L. et al. (2005) Failure to confirm association between RGS4 haplotypes and schizophrenia in Caucasians. Am. J. Med. Genet. B Neuropsychiatr. Genet. 139, 23–27Crossref, Google Scholar
95 Larminie, C. et al. (2004) Selective expression of regulators of G-protein signaling (RGS) in the human central nervous system. Brain Res. Mol. Brain Res. 122, 24–34Crossref, Google Scholar
96 Ross, E.M. and Wilkie, T.M. (2000) GTPase-activating proteins for heterotrimeric G proteins: regulators of G protein signaling (RGS) and RGS-like proteins. Annu. Rev. Biochem. 69, 795–827Crossref, Google Scholar
97 Miyakawa, T. et al. (2003) Conditional calcineurin knockout mice exhibit multiple abnormal behaviors related to schizophrenia. Proc. Natl. Acad. Sci. U. S. A. 100, 8987–8992Crossref, Google Scholar
98 Gerber, D.J. et al. (2003) Evidence for association of schizophrenia with genetic variation in the 8p21.3 gene, PPP3CC, encoding the calcineurin γ subunit. Proc. Natl. Acad. Sci. U. S. A. 100, 8993–8998Crossref, Google Scholar
99 Eastwood, S.L. (2005) Decreased hippocampal expression of the susceptibility gene PPP3CC and other calcineurin subunits in schizophrenia. Biol. Psychiatry 57, 702–710Crossref, Google Scholar
100 Freimer, N. and Sabatti, C. (2004) The use of pedigree, sib-pair and association studies of common diseases for genetic mapping and epidemiology. Nat. Genet. 36, 1045–1051Crossref, Google Scholar