Genetics of Depression
Abstract
Depressive disorders are commonly occurring psychiatric conditions that tend to run in families. They are heterogeneous and complex, predicted to involve the interplay between multiple genes and nongenetic risk factors, such as stressful life events. Their phenomenology and biological substrates are not clearly distinguished from those of normal mood states on the one hand and frequently comorbid conditions such as the anxiety disorders on the other, further complicating research into their etiology. Preliminary molecular genetic studies performed over the past several decades have implicated only a modest number of specific candidate genes in major depression, thus far offering little insight into its pathophysiological basis. This leaves open the way for a new wave of large-scale, genome-wide association studies that are providing vastly increased amounts of preliminary data for understanding these important conditions.
Major depressive disorder (MDD) occurs commonly in the general population. Cross-sectional epidemiological surveys that systematically screen for psychiatric disorders in unselected, representative samples find that approximately 7% of U.S. adults had experienced an episode of MDD sometime in the 12 months before the interview (1). Using retrospective assessment, such surveys estimate that 16% of adults have experienced a major depressive episode during their lifetime; longitudinal studies suggest that cumulative lifetime prevalence rates may be higher still (2, 3). Recurrent MDD, i.e., a condition in which individuals experience multiple clinically significant depressive episodes over the course of their life, occurs in about one-third to two-thirds of these individuals (4); as discussed below, this subtype may have more relevance for genetics. Because of the high prevalence of MDD in countries around the world and the substantial impairment with which it is associated, the World Health Organization has predicted that it would rank second to ischemic heart disease in global burden of disease by 2020 (5). Therefore, understanding its etiological and pathophysiological basis is critical.
This article will review and summarize the extant research regarding the genetics of major depressive disorder. The term “disorder” emphasizes the focus on clinically defined depressive conditions such as MDD rather than a vaguely defined state of “depression.” Some have reasonably argued that the nosological definitions for psychiatric diagnoses, although designed and useful for making clinical assignments and treatment decisions, do not necessarily provide an optimum basis for genetic or other biological lines of inquiry. For example, the precise thresholds requiring a 2-week duration or five of nine associated symptoms for DSM-IV diagnosis of MDD do not seem to represent a genetically distinct condition (6). Nor can we answer the deeper question, with its philosophical and pragmatic implications, as to whether depressive “illness” is categorically different from states of normal sadness (as is assumed in the “medical model”) or simply a more severe manifestation of it on the same continuum of affect. This sort of question has arguably more relevance for depressive disorders than for, say, schizophrenia. If, as the medical model assumes, there are “disease genes” that have specific dysfunction in MDD, then the procedure is to detect these using the classic case-control design, testing for differences in the structure or function of specific genes between those who are affected with and those who are free from the disorder. On the other hand, if the mechanisms underlying normal variation in mood are the same as those that “cause” affective illness, then one could simply study the former in a general population sample to identify the genetic mechanisms underlying MDD. This approach has clear parallels to the study of other common medical conditions such as hypertension and diabetes and their relationship to physiological regulation of blood pressure and glucose homeostasis, respectively. Because of the absence of knowledge of these fundamental features of the entity of clinical depression, I will limit this review to studies that examine the syndrome of MDD, for lack of a better current model.
GENETIC EPIDEMIOLOGICAL STUDIES OF MDD
The usual sequence of establishing the genetic basis of a condition goes from (1) family studies to establish its familial aggregation, to (2) twin and adoption studies to distinguish genetic versus family environmental sources of aggregation, and then to (3) molecular genetic studies to identify specific susceptibility loci and their functional role in producing the condition. Family studies compare rates of illness in relatives of those who have the condition (case probands) with rates in relatives of healthy control subjects. Higher rates in the former group of relatives suggest familial aggregation; i.e., there is an association of illness in probands with illness in their relatives. A systematic meta-analysis that included five well-done family studies of MDD reported a summary odds ratio of 2.84, i.e., individuals biologically related to someone with MDD have almost three times the risk of MDD compared with those without a family history of the disorder (7). To put this statistic into perspective, relatives of patients with schizophrenia have about a 10-fold risk of developing that condition compared with the general population. Both of these are similar to familiar risk estimates for common medical conditions such as type II diabetes but far smaller than those for a Mendelian disorder such as Huntington's disease.
Twin studies allow one to separate genetic from environmental influences by comparing resemblance for the condition within two types of twin pairs: monozygotic twins, who share 100% of their DNA, and dizygotic twins, who share, on average, 50% of their DNA like other sibling pairs. Greater monozygotic than dizygotic resemblance for the condition suggests at least a partial genetic basis. This is quantified by estimating the heritability, the portion of individual differences due to genetic effects. [See Neale and Cardon (8) for more details on twin study methodology.] The meta-analysis cited above includes data from five well-done twin studies of MDD and reports a summary heritability of 37% (7). One large twin study that used longitudinal data to reduce measurement error estimated the reliability-adjusted heritability to be 66% (9). These results suggest that the effects of genes account for one-third to two-thirds of liability to MDD, with the remaining portion of risk due to environmental influences. For the latter, childhood adversity and recent stressful life events are firmly established risk factors in MDD [see, for example, Paykel (10) and Kessler (11)].
Factors affecting genetic risk of MDD
Several modifiers of MDD genetic risk have been identified [reviewed in detail by Levinson (12)]. Recurrent MDD has been shown in some studies to have greater familial risk; thus, having a single depressive episode sometime in one's lifetime may have less to do with genetics than does MDD that recurs. Similarly, for age of onset: more familial forms of the disorder may manifest earlier in life. For these reasons, some molecular genetic researchers seeking to identify depression susceptibility genes have focused their efforts on the phenotype of recurrent, early-onset MDD. Female gender, on the other hand, although an important independent risk factor for MDD (13), has not been reliably associated with an overall difference in familial risk (14), although some of the specific genetic factors may differ between men and women (15). For this reason, some studies stratify their genetic analyses by gender to be sensitive to these effects.
Although too complex to review in detail here, an important consideration for genetic studies of MDD is psychiatric comorbidity. More than half of all individuals with MDD develop an anxiety disorder during their lifetime (16, 17). In addition, many of those who develop MDD or an anxiety disorder have elevated premorbid personality traits such as neuroticism. This observation may be explained, in part, by the suggestion by some genetic epidemiological studies that there may be liability genes that increase risk for internalizing psychopathology in general rather than for specific psychiatric disorders. [For reviews of these studies, see Middeldorp et al. (18) or Hettema (19).] This hypothesis suggests a gene-finding strategy for MDD, adopted by some groups, that blurs diagnostic boundaries in choosing subjects for study, such as including individuals with either MDD or a putatively genetically related condition such as generalized anxiety disorder or simply studying the genetics of neuroticism as a continuous trait in the population.
MOLECULAR GENETIC STUDIES OF MDD
Once a genetic basis for a condition has been demonstrated by genetic epidemiological studies, molecular genetic studies are undertaken to attempt to identify the particular genes underlying that basis. As explained in greater detail by other articles in this issue, the two main types of molecular genetic approaches are linkage and association studies, as applied to MDD below. Before these are reviewed in detail, they need to be placed in their historical context. In classic Mendelian genetic disorders that display specific patterns of familial transmission such as autosomal dominant or recessive, a relatively rare genetic disease strongly aggregates in certain families due to relatively large detrimental effects of a chromosomal abnormality or changes in a specific gene that changes its functional integrity. However, MDD, with complex patterns of genetic transmission similar to those of other common medical conditions such as hypertension or type 2 diabetes, is probably due to the accumulated effects of many genes of small, more subtle effects that interact with each other and environmental factors longitudinally across one's lifespan to “cause” an individual to develop the condition. Many of the early studies, not appreciating this distinction, did not recruit large enough numbers of subjects, making them insufficiently powered to detect genes of small effect. This limitation applies to both linkage and genetic association studies of psychiatric disorders such as MDD, providing one reason among several to explain why it has been difficult to unambiguously identify the liability genes for these conditions.
Linkage studies
Linkage studies, necessarily performed in related individuals such as family pedigrees or sibling pairs, allow one to roughly locate genetic variation underlying a biological trait or condition to particular regions on a chromosome while reducing the likelihood that other regions contain susceptibility loci. In classic Mendelian disorders in which a relatively rare genetic disease strongly aggregated in certain families due to a specific gene, this historically proved to be a powerful technique for localizing disease genes. Using this strategy, several groups performed linkage analyses for MDD, by itself or in combination with related phenotypes, the main findings of which are displayed in Table 1. Few studies found convincing evidence for linkage (genome-wide level significance), and there was little replication across studies for regions with at least suggestive linkage. For MDD, only the short arm of chromosome 1 (1p) contained linkage signals in more than two studies; with this information alone, it is difficult to determine the potential importance of other regions reported.
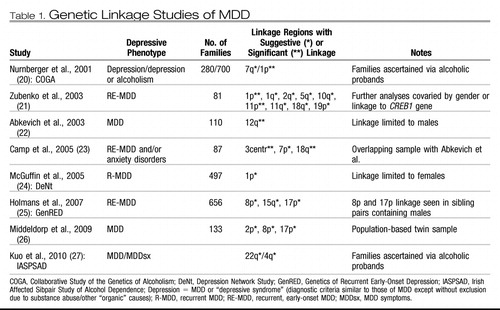 |
Table 1. Genetic Linkage Studies of MDD
Consistent with genetic epidemiological studies that support some anxiety disorders and personality traits such as neuroticism as genetically related to MDD, a number of linkage scans for these other phenotypes have identified genomic regions that overlap those from scans involving MDD, as reviewed in Fullerton (28) and Levinson (12). A meta-analysis of eight independent linkage scans for neuroticism (29) implicated regions on 11q and 12q that overlap with linkage to MDD from Zubenko et al. (21) and Abkevich et al. (22), respectively. As is generally the case with linkage studies, the exact location can be quite variable, and there are typically tens to hundreds of potential candidate genes in the linked regions.
Association studies
Genetic association studies, which may take the form of case-control comparisons in unrelated individuals or family-based transmission tests, allow one to test specific genes or markers within genes for their contribution to normal traits or illness. Most association studies have thus far involved testing one or a few candidate genes implicated indirectly from other data, such as pharmacological agents (e.g., the gene for the serotonin transporter, which underlies the mechanism of action of selective serotonin reuptake inhibitor antidepressants), stress-related biology (e.g., hypothalamic-pituitary-adrenal axis genes), or animal models of depression (e.g., serotonin 1A receptor gene knockouts). The most popularly studied polymorphism in this respect is the 44-base pair insertion/deletion polymorphism (5-HTTLPR) occurring in the promoter region of the serotonin transporter gene (SLC6A4). Both positive and negative reports abound for relating this genetic variant to a myriad of depressive-related phenotypes, making it difficult to draw solid conclusions about its etiological role. The most recent meta-analyses in MDD found a modest main effect for the short (S) allele of this polymorphism (30, 31). Meta-analyses of related phenotypes suggest that there are small but statistically significant effects in association with neuroticism (32–34), suicide (35–37), and obsessive-compulsive disorder (38) but not panic disorder (39). In addition, there is mixed evidence for and subsequent debate over gene-environment interaction effects between 5-HTTLPR and stressful life events (40–42). Such an interaction implies that variation in this gene may increase liability to MDD primarily in individuals who experience significant lifetime stressors such as maltreatment or loss [“genetic control of sensitivity to the environment” (43)].
Similar to linkage studies, most genetic association studies of MDD have been insufficiently powered because of relatively small sample sizes, resulting in a poor record of replicable findings similar to other complex traits (44). Meta-analyses attempt to overcome this limitation by pooling data across studies, effectively increasing the total sample analyzed. However, they are often limited by the inability to include data from all available investigations owing to issues such as heterogeneity between studies or other criteria designed to maximize the quality of the results. Besides the serotonin transporter, separate meta-analyses have examined association of MDD with the gene encoding the dopamine D4 receptor (DRD4) (45), methylenetetrahydrofolate reductase (MTHFR) (46), the serotonin 2A receptor (HTR2A) (47), tyrosine hydroxylase (TH) (48), and angiotensin-converting enzyme (ACE) (49), with only the first two showing significant association across included studies. A recent study reviewed the extant literature for all candidate gene association studies of MDD published before June 2007 (30). This study identified 183 articles that analyzed 393 polymorphisms in 102 candidate genes specifically for association with MDD using acceptable genetic methodology. The authors performed new meta-analyses for 20 polymorphisms in 18 genes that had available data from three or more studies not included in prior meta-analyses. Based on their new analyses together with the prior ones, there is statistical evidence supporting some role for at least five genes in liability to MDD (listed in Table 2): MTHFR, SLC6A3, DRD4, GNB3, and SLC6A4. [Note: Since this analysis, a recent, large study failed to find association with MTHFR (50).] There are probably many more genes involved in MDD, but either (1) insufficient evidence exists for those already studied because of too few or inadequately powered studies or (2) they have yet not been identified as probable candidates for study because of our limited understanding of the pathophysiology of MDD.
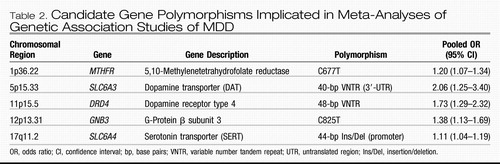 |
Table 2. Candidate Gene Polymorphisms Implicated in Meta-Analyses of Genetic Association Studies of MDD
To overcome the latter limitation, complex genetic conditions are now being studied with genome-wide association studies (GWASs) that use breakthroughs in high-throughput genotyping technology to interrogate upward of hundreds of thousands of polymorphisms across the genome simultaneously in one experiment rather than gene by gene as is done in candidate gene studies. Three GWASs of MDD have been published to date and one is in press. The first, by Muglia et al. (51), reports the findings of GWASs in two large, independent Caucasian samples: (1) 1,022 patients with recurrent MDD and 1,000 matched control subjects recruited from clinical settings in Munich, Germany, and (2) 492 subjects with recurrent MDD and 1,052 healthy control subjects recruited from a community survey in Lausanne, Switzerland. The researchers analyzed about 522,000 single nucleotide polymorphism (SNP) markers in the first sample and about 370,000 in the second. Although the analyses, separately or combined via meta-analysis, did not provide any findings meeting formal genome-wide statistical significance, the authors provided a table of 27 SNPs on 14 chromosomes that provided nominal but consistent evidence for association to MDD across the two samples. Of note, all of these are novel findings; i.e., they are in or near genes that have never been examined before in relation to MDD. The authors also examined association for candidate genes that have been tested in prior mood disorder studies; GRM7, the gene for the metabotrophic glutamate receptor 7 implicated in a recent GWAS of bipolar disorder, emerged as the most significant of these.
Sullivan et al. (52) conducted another large GWAS using the Genetic Association Information Network-MDD (GAIN-MDD) sample: 1,738 Dutch subjects with MDD recruited from clinical and community settings and 1,802 control subjects selected primarily from the Netherlands Twin Registry as having low genetic liability for depressive or anxiety disorders. Similar to the first study, this one also did not identify any associations with genome-wide significance after testing of approximately 435,000 SNPs, and their best signals were from novel candidates, although with no substantial overlap with the top candidates as reported in the first study. They followed up the most convincing candidate gene, PCLO on chromosome 7 coding for the presynaptic protein Piccolo, by testing its association to MDD in five independent samples totaling 6,079 individuals with MDD and 5,893 control subjects. In only one of the replication samples did several of the PCLO SNPs show modest association, although, overall, strict guidelines for replication were not met. However, in a reanalysis of those data Bochdanovits et al. (53) claimed that there is support for a role in liability to MDD for a nonsynonymous coding SNP (rs2522833) in PCLO. Other replication attempts are currently in progress. An independent study performed in a population-based cohort of elderly Dutch individuals recently reported association of rs2522833 with depressive disorders (54).
The third GWAS for MDD (55) was performed in 1,221 Caucasian patients selected from those who participated in the Sequenced Treatment Alternatives to Relieve Depression (STAR*D) (56), a multisite clinical trial; the 1,636 control subjects were drawn from subjects who screened negative for MDD, bipolar disorder, or schizophrenia from the Molecular Genetics of Schizophrenia study in GAIN (57). That analysis, like the others, did not identify any markers with genome-wide significance. Their most strongly associated signals came from a region containing no known genes on 19q12. In that article, the authors combined their data with those from two other MDD GWASs: the Sullivan et al. study in the GAIN-MDD sample described above and an analysis performed in the same 1,636 control subjects from the Molecular Genetics of Schizophrenia sample together with 1,020 subjects from the Genetics of Early-Onset Major Depression (GenRED) sample (listed in Table 1 under linkage analyses) and its follow-up, GenRED II (58). They performed a meta-analysis of these three studies using more than 2.4 million imputed (estimated) SNPs, the most promising of which implicated three genes: (1) ATP6V1B2 on 8p21.3, a gene also modestly implicated in a bipolar disorder GWAS that encodes for a vacuolar proton ATPase, (2) SP4 on 7p15.3, encoding a brain-specific zinc-finger transcription factor, and (3) GRM7 on 3p26.1, the same metabotrophic glutamate receptor gene mentioned in the first MDD GWAS above.
CONCLUSIONS
MDD is a common condition that aggregates in families to a modest degree, most likely due to a large number of genes of small effect. It probably shares some of its genetic risk factors with other internalizing phenotypes such as neuroticism and the anxiety disorders. Linkage studies have not strongly and consistently implicated any specific regions in the genome for harboring susceptibility loci for MDD, a problem that has plagued linkage analyses of other complex disorders. Although hundreds of candidate gene association studies have been published, meta-analyses report only modest evidence supporting five genes thus far: MTHFR (related to folate metabolism), SLC6A3 (dopamine transporter), DRD4 (dopamine receptor type 4), GNB3 (G-protein β subunit 3), and SLC6A4 (serotonin transporter). Thus far, available GWAS data suggest a number of novel susceptibility candidates, including replicated support for the glutamate receptor gene GRM7 gene and, possibly, PCLO; the prior five candidate genes were not among the top-ranked loci. More powerful GWASs, meta-analyses, and replication studies are forthcoming, so this is a rapidly moving target. Except for the serotonin transporter gene, models of gene-environment interaction in MDD, probably an important factor in understanding how genetic factors and life events produce illness in some individuals but not others, have been explored in few studies. Thus, there is still much work to be done before a clear understanding of depression susceptibility genes and their role in the development of MDD will be attained.
1 Kessler RC, Berglund P, Demler O, Jin R, Koretz D, Merikangas KR, Rush AJ, Walters EE, Wang PS: The epidemiology of major depressive disorder: results from the National Comorbidity Survey Replication (NCS-R). JAMA 2003; 289: 3095– 3105Crossref, Google Scholar
2 Moffitt TE, Caspi A, Taylor A, Kokaua J, Milne BJ, Polanczyk G, Poulton, R: How common are common mental disorders? Evidence that lifetime prevalence rates are doubled by prospective versus retrospective ascertainment. Psychol Med 2009; doi: https://doi.org/10.1017/S0033291709991036Google Scholar
3 Patten SB: Accumulation of major depressive episodes over time in a prospective study indicates that retrospectively assessed lifetime prevalence estimates are too low. BMC Psychiatry 2009; 9: 19Crossref, Google Scholar
4 Hardeveld F, Spijker J, De Graaf R, Nolen WA, Beekman AT: Prevalence and predictors of recurrence of major depressive disorder in the adult population. Acta Psychiatr Scand ( in press)Google Scholar
5 Murray CJ, Lopez AD: Alternative projections of mortality and disability by cause 1990–2020: Global Burden of Disease Study. Lancet 1997; 349: 1498– 1504Crossref, Google Scholar
6 Kendler KS, Gardner CO Jr: Boundaries of major depression: an evaluation of DSM-IV criteria. Am J Psychiatry 1998; 155: 172– 177Google Scholar
7 Sullivan PF, Neale MC, Kendler KS: Genetic epidemiology of major depression: review and meta-analysis. Am J Psychiatry 2000; 157: 1552– 1562Crossref, Google Scholar
8 Neale MC, Cardon LR: Methodology for Genetic Studies of Twins and Families. Dordrecht, The Netherlands, Kluwer Academic BV, 1992Google Scholar
9 Kendler KS, Neale MC, Kessler RC, Heath AC, Eaves LJ: The lifetime history of major depression in women. Reliability of diagnosis and heritability. Arch Gen Psychiatry 1993; 50: 863– 870Crossref, Google Scholar
10 Paykel ES: Life events, social support and depression. Acta Psychiatr Scand Suppl 1994; 377: 50– 58Crossref, Google Scholar
11 Kessler RC: The effects of stressful life events on depression. Annu Rev Psychol 1997; 48: 191– 214Crossref, Google Scholar
12 Levinson DF: The genetics of depression: a review. Biol Psychiatry 2006; 60: 84– 92Crossref, Google Scholar
13 Bebbington PE: Sex and depression. Psychol Med 1998; 28: 1– 8Crossref, Google Scholar
14 Merikangas KR, Weissman MM, Pauls DL: Genetic factors in the sex ratio of major depression. Psychol Med 1985; 15: 63– 69Crossref, Google Scholar
15 Kendler KS, Gardner CO, Neale MC, Prescott CA: Genetic risk factors for major depression in men and women: similar or different heritabilities and same or partly distinct genes? Psychol Med 2001; 31: 605– 616Crossref, Google Scholar
16 Kessler RC, Nelson CB, McGonagle KA, Liu J, Swartz M, Blazer DG: Comorbidity of DSM-III-R major depressive disorder in the general population: results from the US National Comorbidity Survey [see comments]. Br J Psychiatry Suppl 1996; 168: 17– 30Google Scholar
17 Regier DA, Rae DS, Narrow WE, Kaelber CT, Schatzberg AF: Prevalence of anxiety disorders and their comorbidity with mood and addictive disorders. Br J Psychiatry Suppl 1998; 173: 24– 28Crossref, Google Scholar
18 Middeldorp CM, Cath DC, van Dyck R, Boomsma D: The co-morbidity of anxiety and depression in the perspective of genetic epidemiology. A review of twin and family studies. Psychol Med 2005; 35: 611– 624Crossref, Google Scholar
19 Hettema JM: What is the genetic relationship between anxiety and depression? Am J Med Genet C Semin Med Genet 2008; 148C: 140– 146Crossref, Google Scholar
20 Nurnberger JI Jr, Foroud T, Flury L, Su J, Meyer ET, Hu K, Crowe R, Edenberg H, Goate A, Bierut L, Reich T, Schuckit M, Reich W: Evidence for a locus on chromosome 1 that influences vulnerability to alcoholism and affective disorder. Am J Psychiatry 2001; 158: 718– 724Crossref, Google Scholar
21 Zubenko GS, Maher B, Hughes HB III, Zubenko WN, Stiffler JS, Kaplan BB, Marazita ML: Genome-wide linkage survey for genetic loci that influence the development of depressive disorders in families with recurrent, early-onset, major depression. Am J Med Genet B Neuropsychiatr Genet 2003; 123: 1– 18Crossref, Google Scholar
22 Abkevich V, Camp NJ, Hensel CH, Neff CD, Russell DL, Hughes DC, Plenk AM, Lowry MR, Richards RL, Carter C, Frech GC, Stone S, Rowe K, Chau CA, Cortado K, Hunt A, Luce K, O'Neil G, Poarch J, Potter J, Poulsen GH, Saxton H, Bernat-Sestak M, Thompson V, Gutin A, Skolnick MH, Shattuck D, Cannon-Albright L: Predisposition locus for major depression at chromosome 12q22–12q23.2. Am J Hum Genet 2003; 73: 1271– 1281Crossref, Google Scholar
23 Camp NJ, Lowry MR, Richards RL, Plenk AM, Carter C, Hensel CH, Abkevich V, Skolnick MH, Shattuck D, Rowe KG, Hughes DC, Cannon-Albright LA: Genome-wide linkage analyses of extended Utah pedigrees identifies loci that influence recurrent, early-onset major depression and anxiety disorders. Am J Med Genet B Neuropsychiatr Genet 2005; 135: 85– 93Crossref, Google Scholar
24 McGuffin P, Knight J, Breen G, Brewster S, Boyd PR, Craddock N, Gill M, Korszun A, Maier W, Middleton L, Mors O, Owen MJ, Perry J, Preisig M, Reich T, Rice J, Rietschel M, Jones L, Sham P, Farmer AE: Whole genome linkage scan of recurrent depressive disorder from the depression network study. Hum Mol Genet 2005; 14: 3337– 3345Crossref, Google Scholar
25 Holmans P, Weissman MM, Zubenko GS, Scheftner WA, Crowe RR, DePaulo JR, Knowles JA, Zubenko WN, Murphy-Eberenz K, Marta DH, Boutelle S, McInnis MG, Adams P, Gladis M, Steele J, Miller EB, Potash JB, MacKinnon DF, Levinson DF: Genetics of recurrent early-onset major depression (GenRED): Final genome scan report. Am J Psychiatry 2007; 164: 248– 258Crossref, Google Scholar
26 Middeldorp CM, Sullivan PF, Wray NR, Hottenga JJ, de Geus EJ, van den Berg M, Montgomery GW, Coventry WL, Statham DJ, Andrews G, Slagboom PE, Boomsma DI, Martin NG: Suggestive linkage on chromosome 2, 8, and 17 for lifetime major depression. Am J Med Genet B Neuropsychiatr Genet 2009; 150B: 352– 358Crossref, Google Scholar
27 Kuo PH, Neale MC, Walsh D, Patterson DG, Riley B, Prescott CA, Kendler KS, Genome-wide linkage scans for major depression in individuals with alcohol dependence. J Psychiatr Res 2010; doi: https://doi.org/10.1016/j.psychires.2009.12.005Google Scholar
28 Fullerton J: New approaches to the genetic analysis of neuroticism and anxiety. Behav Genet 2006; 36: 147– 161Crossref, Google Scholar
29 Hettema JM: Meta-analyses of genome-wide linkage scans of anxiety-related phenotypes, in
30 Lopez-Leon S, Janssens AC, Gonzalez-Zuloeta Ladd AM, Del Favero J, Claes SJ, Oostra BA, van Duijn CM: Meta-analyses of genetic studies on major depressive disorder. Mol Psychiatry 2008; 13: 772– 785Crossref, Google Scholar
31 Kiyohara C, Yoshimasu K: Association between major depressive disorder and a functional polymorphism of the 5-hydroxytryptamine (serotonin) transporter gene: a meta-analysis. Psychiatr Genet ( in press)Google Scholar
32 Sen S, Burmeister M, Ghosh D: Meta-analysis of the association between a serotonin transporter promoter polymorphism (5-HTTLPR) and anxiety-related personality traits. Am J Med Genet B Neuropsychiatr Genet 2004; 127: 85– 89Crossref, Google Scholar
33 Schinka JA, Busch RM, Robichaux-Keene N: A meta-analysis of the association between the serotonin transporter gene polymorphism (5-HTTLPR) and trait anxiety. Mol Psychiatry 2004; 9: 197– 202Crossref, Google Scholar
34 Munafo MR, Freimer NB, Ng W, Ophoff R, Veijola J, Miettunen J, Jarvelin MR, Taanila A, Flint J: 5-HTTLPR genotype and anxiety-related personality traits: a meta-analysis and new data. Am J Med Genet B Neuropsychiatr Genet 2009; 150B: 271– 281Crossref, Google Scholar
35 Anguelova M, Benkelfat C, Turecki G: A systematic review of association studies investigating genes coding for serotonin receptors and the serotonin transporter: II. Suicidal behavior. Mol Psychiatry 2003; 8: 646– 653Crossref, Google Scholar
36 Lin PY, Tsai G: Association between serotonin transporter gene promoter polymorphism and suicide: results of a meta-analysis. Biol Psychiatry 2004; 55: 1023– 1030Crossref, Google Scholar
37 Li D, He L: Meta-analysis supports association between serotonin transporter (5-HTT) and suicidal behavior. Mol Psychiatry 2007; 12: 47– 54Crossref, Google Scholar
38 Lin PY: Meta-analysis of the association of serotonin transporter gene polymorphism with obsessive-compulsive disorder. Prog Neuropsychopharmacol Biol Psychiatry 2007; 31: 683– 689Crossref, Google Scholar
39 Blaya C, Salum GA, Lima MS, Leistner-Segal S, Manfro GG: Lack of association between the serotonin transporter promoter polymorphism (5-HTTLPR) and panic disorder: a systematic review and meta-analysis. Behav Brain Funct 2007; 3: 41Crossref, Google Scholar
40 Caspi A, Sugden K, Moffitt TE, Taylor A, Craig IW, Harrington H, McClay J, Mill J, Martin J, Braithwaite A, Poulton R: Influence of life stress on depression: moderation by a polymorphism in the 5-HTT gene. Science 2003; 301: 386– 389Crossref, Google Scholar
41 Risch N, Herrell R, Lehner T, Liang KY, Eaves L, Hoh J, Griem A, Kovacs M, Ott J, Merikangas KR: Interaction between the serotonin transporter gene (5-HTTLPR), stressful life events, and risk of depression: a meta-analysis. JAMA 2009; 301: 2462– 2471Crossref, Google Scholar
42 Rutter M: Gene-environment interplay. Depress Anxiety 2010; 27: 1– 4Crossref, Google Scholar
43 Kendler KS, Kessler RC, Walters EE, MacLean C, Neale MC, Heath AC, Eaves LJ: Stressful life events, genetic liability, and onset of an episode of major depression in women. Am J Psychiatry 1995; 152: 833– 842Crossref, Google Scholar
44 Hirschhorn JN, Lohmueller K, Byrne E, Hirschhorn K: A comprehensive review of genetic association studies. Genet Med 2002; 4: 45– 61Crossref, Google Scholar
45 Lopez LS, Croes EA, Sayed-Tabatabaei FA, Claes S, Van Broeckhoven C, van Duijn CM: The dopamine D4 receptor gene 48-base-pair-repeat polymorphism and mood disorders: a meta-analysis. Biol Psychiatry 2005; 57: 999– 1003Crossref, Google Scholar
46 Lewis SJ, Lawlor DA, Davey SG, Araya R, Timpson N, Day IN, Ebrahim S: The thermolabile variant of MTHFR is associated with depression in the British Women's Heart and Health Study and a meta-analysis. Mol Psychiatry 2006; 11: 352– 360Crossref, Google Scholar
47 Anguelova M, Benkelfat C, Turecki G: A systematic review of association studies investigating genes coding for serotonin receptors and the serotonin transporter: I. Affective disorders. Mol Psychiatry 2003; 8: 574– 591Crossref, Google Scholar
48 Furlong RA, Rubinsztein JS, Ho L, Walsh C, Coleman TA, Muir WJ, Paykel ES, Blackwood DH, Rubinsztein DC: Analysis and metaanalysis of two polymorphisms within the tyrosine hydroxylase gene in bipolar and unipolar affective disorders. Am J Med Genet 1999; 88: 88– 94Crossref, Google Scholar
49 Lopez-Leon S, Janssens AC, Hofman A, Claes S, Breteler MM, Tiemeier H, van Duijn CM: No association between the angiotensin-converting enzyme gene and major depression: a case-control study and meta-analysis. Psychiatr Genet 2006; 16: 225– 226Crossref, Google Scholar
50 Gaysina D, Cohen S, Craddock N, Farmer A, Hoda F, Korszun A, Owen MJ, Craig IW, McGuffin P: No association with the 5,10-methylenetetrahydrofolate reductase gene and major depressive disorder: results of the depression case control (DeCC) study and a meta-analysis. Am J Med Genet B Neuropsychiatr Genet 2008; 147B: 699– 706Crossref, Google Scholar
51 Muglia P, Tozzi F, Galwey NW, Francks C, Upmanyu R, Kong XQ, Antoniades A, Domenici E, Perry J, Rothen S, Vandeleur CL, Mooser V, Waeber G, Vollenweider P, Preisig M, Lucae S, Muller-Myhsok B, Holsboer F, Middleton LT, Roses AD: Genome-wide association study of recurrent major depressive disorder in two European case-control cohorts. Mol Psychiatry 2008; doi: https://doi.org/10.1038/mp.2008.131Google Scholar
52 Sullivan PF, de Geus EJ, Willemsen G, James MR, Smit JH, Zandbelt T, Arolt V, Baune BT, Blackwood D, Cichon S, Coventry WL, Domschke K, Farmer A, Fava M, Gordon SD, He Q, Heath AC, Heutink P, Holsboer F, Hoogendijk WJ, Hottenga JJ, Hu Y, Kohli M, Lin D, Lucae S, Macintyre DJ, Maier W, McGhee KA, McGuffin P, Montgomery GW, Muir WJ, Nolen WA, Nothen MM, Perlis RH, Pirlo K, Posthuma D, Rietschel M, Rizzu P, Schosser A, Smit AB, Smoller JW, Tzeng JY, van Dyck R, Verhage M, Zitman FG, Martin NG, Wray NR, Boomsma DI, Penninx BW: Genome-wide association for major depressive disorder: a possible role for the presynaptic protein piccolo. Mol Psychiatry 2009; 14: 359– 375Crossref, Google Scholar
53 Bochdanovits Z, Verhage M, Smit AB, de Geus EJ, Posthuma D, Boomsma DI, Penninx BW, Hoogendijk WJ, Heutink P: Joint reanalysis of 29 correlated SNPs supports the role of PCLO/Piccolo as a causal risk factor for major depressive disorder. Mol Psychiatry 2009; 14: 650– 652Crossref, Google Scholar
54 Hek K, Mulder CL, Luijendijk HJ, van Duijn CM, Hofman A, Uitterlinden AG, Tiemeier H: The PCLO gene and depressive disorders: replication in a population-based study. Hum Mol Genet 2010; 19: 731– 734Crossref, Google Scholar
55 Shyn SI, Shi J, Kraft JB, Potash JB, Knowles JA, Weissman MM, Garriock HA, Yokoyama JS, McGrath PJ, Peters EJ, Scheftner WA, Coryell W, Lawson WB, Jancic D, Gejman PV, Sanders AR, Holmans P, Slager SL, Levinson DF, Hamilton SP: Novel loci for major depression identified by genome-wide association study of Sequenced Treatment Alternatives to Relieve Depression and meta-analysis of three studies. Mol Psychiatry 2009; doi: https://doi.org/10.1038/mp.2009.125Google Scholar
56 Rush AJ, Fava M, Wisniewski SR, Lavori PW, Trivedi MH, Sackeim HA, Thase ME, Nierenberg AA, Quitkin FM, Kashner TM, Kupfer DJ, Rosenbaum JF, Alpert J, Stewart JW, McGrath PJ, Biggs MM, Shores-Wilson K, Lebowitz BD, Ritz L, Niederehe G: Sequenced treatment alternatives to relieve depression (STAR*D): rationale and design. Control Clin Trials 2004; 25: 119– 142Crossref, Google Scholar
57 Manolio TA, Rodriguez LL, Brooks L, Abecasis G, Ballinger D, Daly M, Donnelly P, Faraone SV, Frazer K, Gabriel S, Gejman P, Guttmacher A, Harris EL, Insel T, Kelsoe JR, Lander E, McCowin N, Mailman MD, Nabel E, Ostell J, Pugh E, Sherry S, Sullivan PF, Thompson JF, Warram J, Wholley D, Milos PM, Collins FS: New models of collaboration in genome-wide association studies: the Genetic Association Information Network. Nat Genet 2007; 39: 1045– 1051Crossref, Google Scholar
58 Shi J, Potash JB, Knowles JA, Weissman MM, Coryell W, Scheftner WA, Lawson WB, Depaulo JR Jr, Gejman PV, Sanders AR, Johnson JK, Adams P, Chaudhury S, Jancic D, Evgrafov O, Zvinyatskovskiy A, Ertman N, Gladis M, Neimanas K, Goodell M, Hale N, Ney N, Verma R, Mirel D, Holmans P, Levinson DF: Genome-wide association study of recurrent early-onset major depressive disorder. Mol Psychiatry 2010; doi: https://doi.org/10.1038/mp.2009.124Google Scholar