Neuregulin 1 Transcripts Are Differentially Expressed in Schizophrenia and Regulated by 5′ SNPs Associated With the Disease
Abstract
Genetic variation in neuregulin 1 (NRG1) is associated with schizophrenia. The disease-associated SNPs are noncoding, and their functional implications remain unknown. We hypothesized that differential expression of the NRG1 gene explains its association to the disease. We examined four of the disease-associated SNPs that make up the original risk haplotype in the 5′ upstream region of the gene for their effects on mRNA abundance of NRG1 types I–IV in human postmortem hippocampus. Diagnostic comparisons revealed a 34% increase in type I mRNA in schizophrenia and an interaction of diagnosis and genotype (SNP8NRG221132) on this transcript. Of potentially greater interest, a single SNP within the risk haplotype (SNP8NRG243177) and a 22-kb block of this core haplotype are associated with mRNA expression for the novel type IV isoform in patients and controls. Bioinformatic promoter analyses indicate that both SNPs lead to a gain/loss of putative binding sites for three transcription factors, serum response factor, myelin transcription factor-1, and High Mobility Group Box Protein-1. These data implicate variation in isoform expression as a molecular mechanism for the genetic association of NRG1 with schizophrenia.
Schizophrenia is a complex, heritable psychiatric disorder. Recently, several putative schizophrenia susceptibility genes have been identified (1). Genomewide linkage studies and metaanalyses of linkage scans have highlighted chromosome 8p as a susceptibility locus (2–8). Extensive fine-mapping of the 8p locus, haplotype-association analysis, and linkage disequilibrium (LD) tests subsequently implicated neuregulin 1 (NRG1) (6), a gene with pleotropic roles in neurodevelopment and plasticity (9).
The NRG1 gene spans 1.2 Mb (6) and gives rise to many structurally and functionally distinct isoforms, through alternative promoter usage. These isoforms are divided into three classic groups (9): type I (previously known as acetylcholine receptor inducing activity, heregulin, or neu differentiation factor), type II (glia growth factor) and type III (cysteine-rich domain containing), which are based on distinct amino termini. All isoforms have a bio-active EGF-like domain that is responsible for activation of ErbB receptor tyrosine kinases (ErbB2-ErbB4). Additional NRG1 5′ exons have recently been identified, giving rise putatively to novel NRG1 types IV–VI in the human brain (10). No biological information is available presently for these novel isoforms.
In the original report of association with schizophrenia in an Icelandic population, Stefansson and colleagues (6) identified a “core at-risk haplotype” consisting of five SNPs (SNP8NRG221132, SNP8NRG221533, SNP8NRG241930, SNP8NRG243177, and SNP8NRG433E1006) and two microsatellites covering the 5′ end of the NRG1 gene and extending into the second intron (hereafter referred to as the “deCODE haplotype”). Separate follow-up studies in Scottish, Irish, mixed United Kingdom, and Dutch populations confirmed the genetic association between schizophrenia and NRG1 by using markers within the same core haplotype (11–14) or with overlapping markers in the 5′ region (15, 16). Studies in four Asian populations also showed a strong association between schizophrenia and NRG1 polymorphisms at the 5′ (17–20) and 3′ end of the gene (19). Together these results, not withstanding two negative studies (21, 22), provide strong evidence that NRG1 is a schizophrenia-susceptibility gene. Additional support for NRG1’s role in schizophrenia comes from the phenotype of NRG1 and ErbB4 mutant mice (6, 23–25), which exhibit behaviors similar to those of established rodent models of schizophrenia (26).
Exactly how genetic variation in NRG1 impacts on disease susceptibility remains uncertain because the SNPs associated with schizophrenia are all non-coding, being either intronic, synonymous exonic substitutions, or upstream of the transcription start site. It is possible that an as-yet unknown (rare) coding mutation(s) exists, but it is more probable that the noncoding SNPs themselves, or other SNPs with which they are in LD, are functionally associated with the disease. One plausible explanation is that the NRG1 SNPs are regulatory and affect disease susceptibility by altering expression (via altering transcriptional activity, alternative splicing, or stability of the RNA) and, thereby, the amount or distribution of the protein and ultimately its function. Support for this hypothesis comes from data demonstrating increased NRG1 type I mRNA in the prefrontal cortex in schizophrenia (27) and from altered gene expression and splicing being associated with polymorphic variation in other brain diseases (1, 28, 29).
The aim of this study was to address whether disease-associated polymorphic loci in the 5′ upstream region of NRG1 modulate NRG1 mRNA expression and contribute to its association with schizophrenia. We performed a series of hypothesis-driven experiments aimed at confirming previously published positive and negative expression data in schizophrenia (i.e., elevated type I, NRG1, and no change in type II or III (27) and to test specifically the hypothesis that disease-associated SNPs in the original risk haplotype influence expression of the novel type IV isoform based on their physical proximity to its 5′ regulatory region.
To test for differential expression of NRG1 in schizophrenia, we examined mRNA abundance for NRG1 types I–IV in the human hippocampus, a region prominently implicated in the pathogenesis of schizophrenia (30) and in the neurobiology of NRG1 (31). Examination of the effects of genetic variation on NRG1 expression included 4 SNPs from the deCODE core haplotype (6), with each SNP being tested individually for associations with NRG1 mRNA levels in patients and controls. LD between SNPs was examined, and the deCODE at-risk haplotype region was tested for association with NRG1 mRNA abundance. Our results support our primary hypotheses and indicate that the region of the gene implicated by the core at-risk haplotype impacts on specific NRG1 isoforms and interacts with their expression in schizophrenia.
Results
NRG1 Isoform mRNA expression in schizophrenia
Normalized hippocampal NRG1 type I mRNA expression levels were increased by 34% in schizophrenic patients compared with control subjects (Fig. 1; F (1, 85) = 8.65; P = 0.004). Similar significant findings were observed in the less well matched full cohort (84 controls vs. 44 schizophrenics; data not shown). No significant differences were observed between groups for the other NRG1 isoforms {Fig. 1; type II [F (1, 85) = 0.10; P = 0.74]; type III, [F (1, 85) = 0.36; P = 0.54]; type IV, [F (1, 68) = 1.6; P = 0.20}; 17 individuals were not available for the type IV study because of a shortage of RNA]. Expression ratios for all NRG1 isoforms were calculated to investigate relative expression abnormalities, given previous reports of altered isoform ratios in the dorsolateral prefrontal cortex in schizophrenia (27). Type I NRG1 mRNA expression was increased relative to all other isoforms in schizophrenia: type I/II [F (1, 85) = 5.20; P = 0.02], I/III [F (1, 85) 5.0; P = 0.02], and I/IV [F (1, 68) = 10.11; P = 0.002]. No changes in any other isoform ratio were seen. No significant differences were observed between diagnostic groups for any of the housekeeping genes PBGD, [F (1, 85) = 0.27; P = 0.6]; TBP, [F (1, 85) = 0.16; P = 0.21], and GUSB, [F (1, 68) = 0.001; P = 0.98]. Covariation for pH, postmortem interval (PMI), and age was used in all analyses (see Supporting Text, which is published as supporting information on the PNAS web site).
Effects of 5′ SNPs on NRG1 expression
The effect of SNPs on NRG1 type I–III isoform expression were examined in the whole cohort (n = 84 controls and n = 44 schizophrenics). For type IV analysis, 24 individuals were not available for analysis because of a shortage of RNA (leaving n = 74 controls and n = 30 schizophrenics). None of the SNPs examined showed any effect on expression of type II or type III isoforms.
Genetic variation and type I NRG1 mRNA
A genotype × diagnosis interaction [F (3, 122) = 9.21; P = 0.003] was found for SNP8NRG221132 and type I NRG1 mRNA. There was no main effect of genotype. Post hoc comparisons showed that the genotype was significant in the controls alone (post hoc t test; P = 0.003; Fig. 2) with individuals carrying the rare A (2) allele expressing higher levels than individuals homozygous for G (1/1 genotype). The G allele constitutes the risk allele in the deCODE haplotype (6, 11). However, this pattern was not seen in schizophrenic patients who tended in the opposite direction (Fig. 2). Schizophrenic patients homozygous for the risk allele (G) had increased expression of type I NRG1 mRNA compared with control subjects homozygous for the same allele (t test; P = 0.001). To confirm these findings, we genotyped SNP8NRG221132 in brain tissue from an earlier study of a separate cohort, in whom NRG1 mRNA expression for types I–III had been measured by identical quantitative RT-PCR methods (controls n = 13, schizophrenics n = 16; n= 22 African Americans and n = 7 Caucasians) (27), and NRG1 type I expression was found to be increased in the dorsolateral prefrontal cortex of these patients with schizophrenia. In this cohort we observed a main effect of SNP8NRG221132 genotype in the whole sample [F (3, 25) = 15.17; P = 0.0005] on pre-frontal type I NRG1 mRNA, with subjects carrying the A allele (n = 4) again showing higher type I NRG1 levels compared with homozygous G cases (n = 25, data not shown). A genotype × diagnosis interaction also was found in this cohort [F (3, 25) = 16.26; P = 0.0005], with schizophrenic patients homozygous for the G allele (n = 14) having greater expression of type I NRG1 mRNA than control subjects (n = 11) with the same genotype (P = 0.02). No other SNPs examined in the study were associated with type I NRG1 expression.
Genetic variation and type IV NRG1 mRNA
Because >10% of individuals in the cohort were homozygous for the rare risk allele (T) at SNP8NRG243177, a complete analysis was conducted based on the three genotype groups (C/C, C/T, and T/T). We found a main effect of genotype for SNP8NRG243177 on type IV NRG1 mRNA abundance in the whole sample [F (5, 98) = 3.15; P = 0.04; Fig. 3A]. The data suggest an allele dose effect with individuals heterozygous for the (T) risk allele (6, 11) having 21% more type IV NRG1 mRNA than homozygous C/C individuals and individuals homozygous for the risk allele having 49% more type IV NRG1 mRNA expression than homozygous C/C individuals. However, standard post hoc contrasts between the three groups revealed only the two homozygote groups to be significantly different in terms of type IV abundance (post hoc t test; P = 0.05; Fig. 3A). This effect appeared more pronounced in the schizophrenia group alone (Fig. 3B), although no hint of a diagnosis × genotype interaction was observed. Of note, normal control individuals homozygous for the T allele also showed the relatively greatest type IV NRG1 mRNA expression (Fig. 3B).
Significant diagnosis × genotype interactions were found between five of the six haplotype-tagging SNPs (htSNPs) and type IV mRNA abundance (rs4268090, rs4298458, rs4452759, rs4733263, and rs4476964) [range F (3, 100) = 3.35–5.82; P = 0.033–0.018]. A trend for a main effect of genotype was observed for the htSNP, rs4268090 [F (3, 100) = 3.55; P = 0.06]. Carrying the T (2) allele at this SNP was associated with higher levels of type IV mRNA compared with subjects homozygous for the C (1/1) allele.
Haplotype analysis
Results of LD tests between pairs of all 10 5′ SNPs in African American and Caucasian individuals can be found in Tables 1 and 2, which are published as supporting material on the PNAS web site. The four markers chosen from the deCODE haplotype were in significant, but moderate, LD in both groups (see Supporting Text). The frequencies for the four common haplotypes comprised of the four deCODE SNPs are shown in Table 3, which is published as supporting information on the PNAS web site). Hap 4 contains the specific alleles that form part of the deCODE haplotype.
To test whether this four SNP risk haplotype (hap4) was associated with NRG1 mRNA levels, we used snphap (wwwgene.cimr.cam.ac.uk/clayton/software) to assign a diplotype (haplotype pair) to each individual. We then compared hap4 carriers (diplotypes hap1/hap4, hap2/hap4, hap3/hap4, and hap4/hap4) to non-hap4 individuals (diplotypes hap1/hap1, hap1/hap2, hap1/hap3, hap2/hap2, hap3/hap3, and hap2/hap3), testing for an effect of carrying the risk haplotype on NRG1 mRNA abundance. ANOVA revealed a main effect of hap4 on type IV mRNA abundance in the entire sample [F (3, 89) = 3.38; P = 0.04]. Hap4 carriers had 27% more type IV mRNA compared with non-hap4 individuals (Fig. 4A). This effect appeared more pronounced in the schizophrenic patients, where a 53% increase in type IV NRG1 mRNA was seen in hap4 carriers compared to noncarriers; in controls, only a 14% increase was observed (Fig. 4B). However, no diagnosis by genotype interaction was found. Race was not included as a factor in the analysis because of the small number of African American individuals carrying hap4. Hap4 showed no effect on the expression of any of the other NRG1 isoforms.
Promoter analysis based on transcription factor binding sites
As a further exploration of the functional relevance of disease-associated SNPs in the NRG1 gene, we performed an analysis of putative transcription factor binding sites by using matinspector software (Genomatix, Munich), a computational suite for promoter informatics. Two SNPs in the haplotype were indicated to be contained within transcriptional regulatory elements; notably, these elements were the two SNPs that, as described above, were found to impact upon expression of NRG1 isoforms, SNP8NRG221132, which is associated with type I NRG1, and SNP8NRG243177, which is associated with type IV NRG1. SNP8NRG221132 is within a predicted transcription factor binding domain for serum response factor (SRF), with the risk allele (G) abolishing SRF binding. SNP8NRG243177 is also within a putative binding site for SRF and for myelin transcription factor 1. Carrying the risk allele (T) results in a predicted loss of binding to both of these transcription factors and the acquisition of the transcription factor binding site for High Mobility Group Box Protein-1. None of the other 8 SNPs genotyped in the study mapped to transcription factor binding domains.
Analysis of negative SNP controls
The two negative control SNPs (rs10954867 and rs7005288) showed no association with any NRG1 isoform in either controls or schizophrenic patients (all P > 0.2).
Regional distribution of hippocampal NRG1 mRNA IN schizophrenia
Because no information is available regarding the distribution of NRG1 in the human hippocampus, and this data was not provided from the quantitative RT-PCR experiments, we examined NRG1 mRNA in the hippocampus in schizophrenia by using in situ hybridization with a “pan” NRG1 probe (Supporting Text; see also Fig. 5, which is published as supporting information on the PNAS web site). No differences were seen in the distribution of pan NRG1 mRNA between subfields or its overall abundance in schizophrenia.
Discussion
We have investigated the expression of NRG1 type I–IV mRNA in the human hippocampus and examined the effects of schizophrenia and disease-associated polymorphisms in the 5′ upstream region on expression of these transcripts. We hypothesized that the genetic association of NRG1 with schizophrenia is mediated by altered expression of the gene based on the location and noncoding nature of the disease-associated polymorphisms and the fact that extensive sequencing of NRG1 has failed to identify pathogenic coding mutations (6). We report three principal findings: (i) up-regulation of type I expression in the hippocampus in schizophrenia, (ii) association of type I expression with a single SNP residing in the original deCODE risk haplotype, and (iii) association of type IV expression with a single SNP and a four-marker haplotype representing the 5′ upstream region of the original at-risk haplotype associated with schizophrenia. We provide evidence of association between disease linked-variation in NRG1 and altered NRG1 isoform expression in the brain, and we propose that altered transcript regulation is a potential molecular mechanism behind the genetic association of NRG1 with schizophrenia.
Our finding of increased type I mRNA NRG1 expression in the hippocampus in schizophrenia replicates the finding in the dorsolateral prefrontal cortex of a smaller and separate brain series (27). These findings suggest that enhanced type I expression is robust and found in two separate brain regions in schizophrenia. In addition, we also replicate the finding that type II and type III isoform expression is unaltered in schizophrenia, suggesting that these isoforms may not be directly relevant to the pathophysiology of the disease. However, we did observe increases in the relative abundance of type I to type II–IV, suggesting that the contribution of these latter isoforms to NRG1 signaling in the hippocampus may be indirectly compromised in patients with schizophrenia. At present, it is unclear whether type I up-regulation in schizophrenia is primary or secondary to other abnormalities in NRG1 isoform regulation or to other molecular changes associated with the disease.
When the four individual SNPs representing the 5′ region of the deCODE at-risk haplotype were tested for association with type I NRG1 mRNA, no main effects of genotype were seen for any of the SNPs. A diagnosis × genotype interaction was observed at a single SNP, SNP8NRG221132, and post hoc tests showed an effect of genotype only in control subjects on type I mRNA abundance. In a second independent cohort of brains in which increased type I mRNA expression previously had been reported in the schizophrenia samples, we found a main effect of a genotype at this SNP in the entire sample and again a genotype by diagnosis interaction. This main effect of the genotype was not seen in the first cohort; however, we note that the main effect in the entire sample is driven primarily by the controls. The observations that the main effect of the genotype was primarily in the controls, and that the four-marker risk haplotype had no effect on type I NRG1 expression, raise the possibility that SNP8NRG221132 influences type I expression independent of its contribution to risk for schizophrenia. Additional support for the functional relevance of SNP8NRG221132 comes from the bioinformatic promoter analysis that predicts the risk allele (G) leads to a loss of binding for SRF. SRF is a transcription factor that regulates the expression of genes encoding cytoskeletal proteins, such as cofilin and actin (32), both of which have been linked directly to NRG1s role in actin dynamics (33). The loss of SRF binding in controls homozygous for the risk allele, therefore, may be related to lower levels of type I mRNA transcription, as reported here. The direct functional consequences of this SNP for type I NRG1 transcriptional control remain difficult to predict because the SNP resides 1 Mb upstream from the transcriptional start site of type I. However, SNP8NRG221132 could conceivably reside in a regulatory element of the gene, as is seen in other key developmental genes where genomic regions harboring cisregulatory elements can be located as far as 1 Mb from the transcription unit (34).
The interrelationship between SNP8NRG221132, type I NRG1 expression, and schizophrenia is somewhat more difficult to interpret, because in contrast to the effect seen in controls, we did not see a similar genotype effect in the patients. This issue is discussed in Supporting Text, Discussion: Genetic Association and Type I Expression in Schizophrenia).
In contrast to the type I finding, which is not manifestly related to genetic variation in NRG1 associated with schizophrenia, the association between both SNP8NRG243177 and the four-marker at-risk haplotype with expression of a novel isoform of NRG1, type IV, suggests that we may have identified a genetic mechanism and a molecular phenotype underlying the involvement of NRG1 in susceptibility for schizophrenia. The risk allele of SNP8NRG243177 and the deCODE haplotype predicted higher levels of type IV NRG1 expression in our entire sample. Analysis of the three genotype groups for SNP8NRG243177 revealed that individuals homozygous for the risk allele had the highest levels of type IV expression, with evidence of an allele dose-dependant effect. This observation appeared more pronounced in the patients, although trends in the same direction were found in the normal controls and no diagnosis × genotype interaction was observed. SNP8NRG243177 is the most 3′ of the SNPs in the four-marker deCODE haplotype and is located ≈1.2 kb upstream of the transcriptional start of type IV. Because none of the other single SNPs in this haplotype were associated with type IV NRG1 expression, our results suggest that SNP8NRG243177 is a functional polymorphic variant that regulates type IV NRG1 mRNA levels or is in strong LD with a nearby functional mutation. Additional support for the functional relevance of SNP8NRG243177 for gene regulation comes from the bioinformatic prediction that this SNP determines a putative transcription factor binding domain for SRF, myelin transcription factor 1, and High Mobility Group Box Protein-1. Of note, SRF and myelin transcription factor 1 play critical roles in neuronal migration, synaptic plasticity, and oligodendrocyte proliferation and survival, respectively, providing a striking molecular convergence with current hypotheses regarding the neurobiology of schizophrenia and the potential role of NRG1 (35). However, we do not know which, if any, of these changes in transcription factor binding sites might mediate the association between SNP8NRG243177 and type IV expression and schizophrenia. Of potential interest is High Mobility Group Box Protein-1, an abundant chromatin-binding protein, which acts as an architectural facilitator in transcription (36). In our sample, acquisition of two High Mobility Group Box Protein-1-binding motifs (i.e., homozygosity for the risk allele) was associated with significantly elevated type IV NRG1 expression, whereas acquisition of one (i.e., heterozygosity for the risk allele) was not. This observation suggests (i) that this binding site may potentiate type IV NRG1 transcription (and that SRF binding is necessary for optimal levels of type IV transcription) and 2) that this effect may be recessive.
The absence of overall changes in type IV NRG1 gene expression levels in schizophrenia suggest that altered type IV, unlike type I, is not a general characteristic of the disease state, per se. Indeed, if altered NRG1 type IV expression is part of the genetic architecture of susceptibility for schizophrenia, it would not be expected to show an effect at the general population level, assuming that the at-risk haplotype is relevant for, at most, 10% of cases. Furthermore, our finding that the deCODE risk haplotype is associated specifically with type IV NRG1 expression argues that the clinical association with NRG1 is based on this molecular effect.
We further report association of type IV NRG1 mRNA in schizophrenia with five additional htSNPs, which span a 17-kb gap between the four SNPs from the deCODE haplotype. To our knowledge, these SNPs have not been tested for association with schizophrenia in the same clinical samples in which the deCODE SNPs were positive. We genotyped these SNPs to address the possibility that the deCODE haplotype might not provide sufficient information regarding genetic diversity in our sample. None of these SNPs showed main effects, and their association with NRG1 type IV expression in schizophrenia is likely via LD with SNP8NRG243177.
In our sample, the deCODE risk haplotype, which we termed hap4, was present in both Caucasian and African American populations but more common in the Caucasian sample. The significant degree of LD across this region of the gene suggests that, at least in Caucasians, it has undergone very little recombination (37). Furthermore, the region is highly conserved between species, including chimpanzee, dog, mouse, and rat, suggesting that this region of the gene is functional, probably involved in transcriptional regulation of NRG1 (38). We found no evidence to suggest that the frequency of the deCODE haplotype was higher in our patient population compared with controls, but our sample is too small to meaningfully test for association with clinical phenotype. Of note, we observed that the frequency of hap2 was somewhat greater in the African American patients (34%) compared with African American controls (25%), suggesting that in different ethnic groups, different haplotypes in the same region of the gene may be associated with schizophrenia. However, because of the small sample size involved, conclusions are limited. Interestingly, hap2 in the African American sample contains the same allele at SNP8NRG243177 as hap4.
In the original report by Stefansson et al. (6), association in Icelandic families was mapped to a seven-marker haplotype spanning a 270-kb LD block starting at SNP8NRG221132 and ending with a synonymous SNP in exon two (SNP8NRG433E1006) and two microsatellites in the second intron (478N14-848 and 420M9-13950). Evidence of association to this region of the gene in other samples has been primarily to SNPs at the 5′ end of this haplotype, encompassing the SNPs typed in this study. Thus, although we cannot exclude the possibility that the causative mutation(s) accounting for our association with type IV lies downstream from our typed SNPs, we tend to doubt this possibility for three reasons: (i) the exon 2 SNP and the microsatellites typed in the deCODE haplotype have not shown single point (pairwise) association with schizophrenia in any study (6, 18, 19, 39), in contrast to the four SNPs tested here, (ii) the physical location of SNP8NRG243177 (i.e., ≈1,200 bases upstream from the exon 1 start site) makes it a far better candidate for being located in a transcriptional regulatory region for type IV, and (ii) this SNP is in a putative functional transcription factor binding domain.
The known biological functions of NRG1 (9) fit well with current hypotheses regarding the neurobiology of schizophrenia (35), including regulation of synaptogenesis, in vivo synaptic transmission, long-term potentiation, activity-dependent synaptic plasticity, and neuronal migration as well as neurotransmitter function (NMDA, GABA, α-7, and dopamine) and oligodendrocyte biology, all of which are proposed to interact or be altered in schizophrenia (30, 40). Of particular relevance is the recent finding that NRG1 down-regulates NMDA-receptor currents in prefrontal cortical pyramidal neurons and slices (41). These data suggest that increased expression of NRG1 type I or IV would translate into decreased NMDA receptor-mediated signaling, one of the principal neurotransmitter hypotheses of schizophrenia.
Finally, it should be noted that we have performed a number of tests in this study, and correction for multiple testing was not performed. Correction for random effects, such as Bonferroni correction, would be an excessively conservative approach, particularly given that we have restricted our primary analyses to planned comparisons (based on strong prior clinical association and physical location of the SNPs) of four SNPs and a single haplotype comprised of these SNPs. Because the SNPs are in moderate LD, the degree of independence between markers is low and, therefore, correcting for multiple testing would result in a high type II error rate. The prior probability and the predictable association between the deCODE haplotype and expression of NRG1 isoforms (especially type IV, which is its immediate physical neighbor) combined with the LD between SNPs in this haplotype makes statistical correction for these comparisons inappropriate. Nevertheless, our finding regarding type IV expression and the deCODE haplotype and SNP8NRG243177 requires independent replication.
In summary, we provide evidence of splice variant-specific alterations of NRG1 gene expression in schizophrenia and demonstrate that disease-associated polymorphisms in a 5′ regulatory region of NRG1 are associated with differential NRG1 isoform expression. We suggest that the mechanism behind the clinical association of NRG1 with schizophrenia is altered transcriptional regulation, which modifies, probably to a small degree and in an isoform-limited fashion, the efficiency of NRG1 signaling effects on neural development and plasticity. Such alterations may compromise cortical and hippocampal function through one or more of the roles of NRG1 and reflect, at least partly, the contribution of NRG1 to the genetic risk architecture for the disease.
Materials and methods
Human postmortem tissue
Postmortem hippocampal tissue was collected at the Clinical Brain Disorders Branch, National Institute of Mental Health, from 84 normal controls (22 females/62 males, 53 African American/25 American Caucasian/5 Hispanic/1 Asian, mean age 40.5 ± (SD) 15.4 years, PMI 30.7 ± 13.9 h, pH 6.59 ± 0.32) and 44 schizophrenic patients (15 females/29 males, 24 African Americans/20 Caucasians, mean age 49.7 ± 17.2 years, PMI 36.3 ± 17.7 h, pH 6.48 ± 0.28). This whole cohort was used for the analysis of effects of genetic variation on NRG1 isoform expression. The different genotype groups in this cohort did not differ on any of the potential variables that affect gene expression in the human postmortem brain (i.e., age, PMI, pH, and age). Because the diagnostic groups in the whole cohort were not perfectly matched for these variables, we selected a subcohort of 53 controls (17 females/36 males, 31 African Americans/17 Caucasians/5 Hispanic individuals, mean age 44 ± 14.2 years, PMI 33.3 ± 13.7, pH 6.53 ± 0.24) and 38 schizophrenic individuals (12 females/26 males, 18 African Americans/19 Caucasians/1 Hispanic individuals, mean age 49.3 ± 19.3 years, PMI 38.1 ± 18.8, pH 6.40 ± 0.26), matched for these potential confounding variables. This subcohort was used for diagnostic comparisons of NRG1 expression levels. Details of brain collection, neuroleptic medication history, and RNA extraction are described in Supporting Text.
Oligonucleotide and primer design
Primer and probe designs for NRG1 types I–III were as described in ref. 27. Details of type IV and type I–III design can be found in Supporting Text (see also Table 4, which is published as supporting information on PNAS web site).
Quantitative real-time RT-PCR
NRG1 mRNA expression levels were measured by quantitative RT-PCR by using an ABI Prism 7900 sequence detection system with a 384-well format (Applied Biosystems) as described (see Supporting Text). Our primary data analysis is based on normalization of NRG1 mRNAs to an endogenous control gene, because it accounts for variability in the initial concentration and quality of total RNA and in the conversion efficiency of the reverse transcription reaction (42). Optimal normalization of a target mRNA to an endogenous control gene requires that the two transcripts have similar expression levels and that the control gene expression levels do not differ between the comparison groups. Based on this finding, PBGD was considered the most reliable for normalization here. Similar results were, however, obtained with normalization to TBP or GUSB (data not shown).
NRG1 genotype determination
DNA was extracted from cerebellar tissue by using a standard protocol supplied by PUREGENE (Gentra Systems). We genotyped four SNPS from the de-CODE core haplotype (SNP8NRG221132, SNP8NRG221533, SNP8NRG241930, and SNP8NRG243177) and selected six additional SNPs (rs10096573, rs4268090, rs4298458, rs4452759, rs4733263, and rs4476964; Table 5 and Fig. 6, which are published as supporting material on the PNAS web site) from HAPMAP (www.hapmap.org) based on designation of these as htSNPS by using HAPLOVIEW (www.broad.mit.edu/mpg/haploview). The additional markers define the common haplotypes in an LD block containing a 22-kb region upstream of the first exon in type IV NRG1, which includes the four most 5′ SNPs of the deCODE core risk haplotype. These six htSNPs were chosen to maximize genetic coverage because they are highly informative tags for the common haplotypes in this region of the gene.
Two SNPs at the 3′ end of NRG1 were selected from the dbSNP database as negative control genotypes (rs10954867 and rs7005288). These SNPs previously have not been associated with schizophrenia, are not in known regulatory domains, and were included in the analysis as a control for random statistical effects. Genotyping was performed by using the Taqman 5′ exonuclease allelic discrimination assay (details available upon request). Genotype reproducibility was routinely assessed by re-genotyping all samples for selected SNPs and was generally >99%. LD between 5′ SNPs was determined by using the program ldmax/gold (43). The program snphap written by David Clayton (version 1.0) was used to calculate haplotype frequencies and to assign diplotypes to individuals.
Statistical analyses
Correlations of mRNA levels with demographic variables were performed for all subjects by using Spearman’s correlations. Correlations of mRNA levels with neuroleptic medication (lifetime neuroleptic exposure, daily dose, and final neuroleptic dose) were investigated in the schizophrenic cohort. Primary planned comparisons between diagnostic groups were made by using univariate ANCOVA for each mRNA with diagnosis as the independent variable and age, pH, and PMI as covariates. Effects of genetic variation on NRG1 mRNA expression were examined by using ANOVA with genotype and diagnosis as independent factors. Primary comparisons examined the effects of four SNPs and the core haplotype on type I–IV expression in patients and controls. Secondary, post hoc analyses included examination of the 6 htSNPS, where warranted. Where there was a significant genotype × diagnosis interaction, individual group post hoc tests were examined as part of the standard ANOVA readout. Analysis of the effects of race was restricted to African American and Caucasian individuals because of the small sample size in other ethnic groups. The genotype groups did not differ on any of the demographic variables, and no correlations were seen between NRG1 isoform expression, age, pH, or PMI in the different genotype groups; therefore straight ANOVAs are reported for the effects of genotype, but the statistical results were not changed when covariates were included. To increase power for statistical analyses of SNPs with minor allele frequencies <10%, we grouped individuals heterozygous and homozygous for the rare allele. Examination of all three genotype groups was conducted when the minor allele frequency was >10%. All experiments were conducted blind to diagnosis.
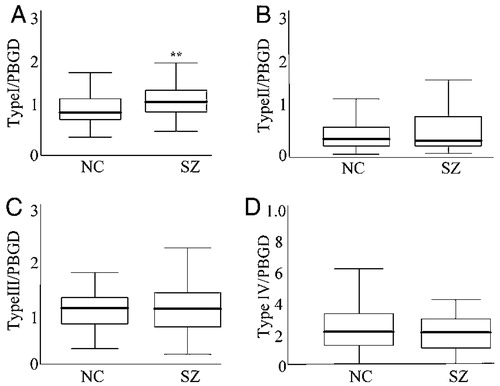
Figure 1. NRG1 types I–IV mRNA expression in the hippocampus of schizophrenic patients and control individuals. Quantitative RT-PCR analysis of NRG1 types I–IV normalized to PBGD. (A–C) n = 53 normal control subjects (NC) and 38 patients with schizophrenia (SZ). (D) n = 46 normal controls subjects and 28 patients with schizophrenia. Box represents the proportion of the distribution falling between the 25th and 75th percentiles. Bars outside the box represent the SD. Bar inside represents the mean. Significant differences were found between controls and patients for type I NRG1 expression. **, significant differences (P < 0.01).
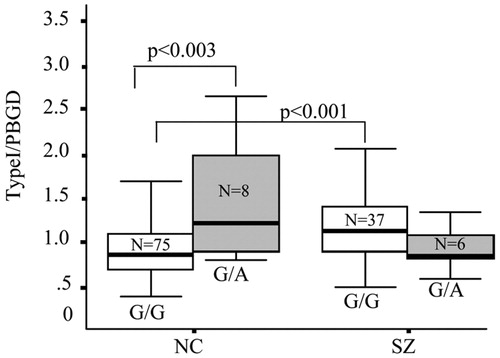
Figure 2. Association between SNP8NRG122132 and type I NRG1 mRNA in normal controls (NC) and schizophrenics (SZ). A significant interaction of genotype and diagnosis was observed on normalized type I mRNA expression (P < 0.003). Post hoc t tests revealed a main effect of genotype on type I NRG1 expression in controls, with A allele carriers having increased levels compared with homozygous G individuals (P < 0.003). Schizophrenic patients homozygous for the G allele had higher levels of type I mRNA compared with controls with the same genotype (P < 0.001). Two individuals were excluded from analysis because of genotyping failure. Box represents the proportion of the distribution falling between the 25th and 75th percentiles. Bars outside the box represent the SD. The bar inside represents the mean.
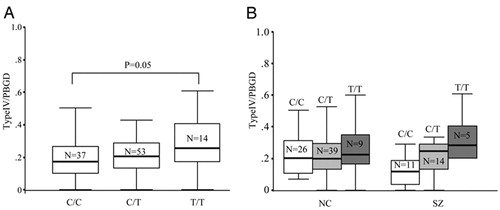
Figure 3. Association between SNP8NRG-243177 and type IV NRG1 mRNA expression. (A) In the whole cohort, a main effect of genotype was observed (ANOVA; P = 0.04) An allele dose-dependent effect is suggested, with individuals homozygous for the risk allele having the highest levels of type IV NRG1 mRNA (P = 0.05). (B) Data parsed by diagnosis. No genotype × diagnosis interaction was observed.
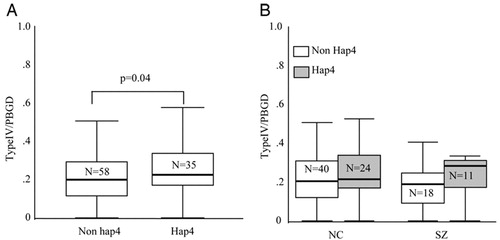
Figure 4. Association between diplotypes containing the deCODE risk haplotype (hap4) and type IV NRG1 mRNA. Individuals were divided according to diplotype into two groups, nonhap4 carriers (haplotypes 1/1; 1/2, 1/3, 2/2, 3/3, and 2/3) and hap4 carriers (haplotypes 1/4, 2/4, 3/4, and 4/4). A main effect of diplotype was observed on normalized type IV NRG1 mRNA levels (ANOVA; P = 0.04). (A) Individuals carrying the hap4 risk haplotype had increased levels compared with individuals not carrying hap4. (B) Effect of diplotype on type IV NRG1 mRNA levels in controls and patients. Eleven individuals were not included in the diplotype analysis because of either the failure of genotyping at one or more of the SNPs or low probability (<93%) of diplotype assignment according to snphap.
1 Harrison, P. J. & Weinberger, D. R. (2005) Mol. Psychiatry 10, 40–68.Crossref, Google Scholar
2 Pulver, A. E., Lasseter, V. K., Kasch, L., Wolyniec, P., Nestadt, G., Blouin, J. L., Kimberland, M., Babb, R., Vourlis, S., Chen, H., et al. (1995) Am. J. Med. Genet. 60, 252–260.Crossref, Google Scholar
3 Kendler, K. S., MacLean, C. J., O’Neill, F. A., Burke, J., Murphy, B., Duke, F., Shinkwin, R., Easter, S. M., Webb, B. T., Zhang, J., et al. (1996) Am. J. Psychiatry 153, 1534–1540.Crossref, Google Scholar
4 Kendler, K. S., Myers, J. M., O’Neill, F. A., Martin, R., Murphy, B., MacLean, C. J., Walsh, D. & Straub, R. E. (2000) Am. J. Psychiatry 157, 402–408.Crossref, Google Scholar
5 Blouin, J. L., Dombroski, B. A., Nath, S. K., Lasseter, V. K., Wolyniec, P. S., Nestadt, G., Thornquist, M., Ullrich, G., McGrath, J., Kasch, L., et al. (1998) Nat. Genet. 20, 70–73.Crossref, Google Scholar
6 Stefansson, H., Sigurdsson, E., Steinthorsdottir, V., Bjornsdottir, S., Sigmundsson, T., Ghosh, S., Brynjolfsson, J., Gunnarsdottir, S., Ivarsson, O., Chou, T. T., et al. (2002) Am. J. Hum. Genet. 71, 877–892.Crossref, Google Scholar
7 Badner J. A. & Gershon E. S. (2002) Mol. Psychiatry 7, 405–411.Crossref, Google Scholar
8 Lewis, C. M., Levinson, D. F., Wise, L. H., DeLisi, L. E., Straub, R. E., Hovatta, I., Williams, N. M., Schwab, S. G., Pulver, A. E., Faraone, S. V., et al. (2003) Am. J. Hum. Genet. 73, 34–48.Crossref, Google Scholar
9 Falls, D. L. (2003) J. Neurocytol. 32, 619–647.Crossref, Google Scholar
10 Steinthorsdottir, V., Stefansson, H., Ghosh, S., Birgisdottir, B., Bjornsdottir, S., Fasquel, A. C., Olafsson, O., Stefansson, K. & Gulcher, J. R. (2004) Gene 342, 97–105.Crossref, Google Scholar
11 Stefansson, H., Sarginson, J., Kong, A., Yates, P., Steinthorsdottir, V., Gudfinnsson, E., Gunnarsdottir, S., Walker, N., Petursson, H., Crombie, C., et al. (2003) Am. J. Hum. Genet. 72, 83–87.Crossref, Google Scholar
12 Williams, N. M., Preece, A., Spurlock, G., Norton, N., Williams, H. J., Zammit, S., O’Donovan, M. C. & Owen, M. J. (2003) Mol. Psychiatry 8, 485–487.Crossref, Google Scholar
13 Bakker, S. C., Hoogendoorn, M. L., Selten, J. P., Verduijn, W., Pearson, P. L., Sinke, R. J. & Kahn, R. S. (2004) Mol. Psychiatry 9, 1061–1063.Crossref, Google Scholar
14 Hall, D., Gogos, J. A., Karayiorgou, M. (2004) Genes Brain Behav. 3, 240–248.Crossref, Google Scholar
15 Corvin, A. P., Morris, D. W., McGhee, K., Schwaiger, S., Scully, P., Quinn, J., Meagher, D., Clair, D. S., Waddington, J. L. & Gill, M. (2004) Mol. Psychiatry 9, 208–213.Crossref, Google Scholar
16 Petryshen, T. L., Middleton, F. A., Kirby, A., Aldinger, K. A., Purcell, S., Tahl, A. R., Morley, C. P., McGann, L., Gentile, K. L., Rockwell, G. N., et al. (2005) Mol. Psychiatry 10, 366–374.Crossref, Google Scholar
17 Yang, J. Z., Si, T. M., Ruan, Y., Ling, Y. S., Han, Y. H., Wang, X. L., Zhou, M., Zhang, H. Y., Kong, Q. M. & Liu, C. (2003) Mol. Psychiatry 8, 706–709.Crossref, Google Scholar
18 Tang, J. X., Chen, W. Y., He, G., Zhou, J., Gu, N. F., Feng, G. Y. & He, L. (2004) Mol. Psychiatry 1, 11–20.Google Scholar
19 Li, T., Stefansson, H., Gudfinnsson, E., Cai, G., Liu, X., Murray, R. M., Steinthorsdottir, V., Januel, D., Gudnadottir, V. G., Petursson, H., Ingason, A., et al. (2004) Mol. Psychiatry 9, 698–704.Crossref, Google Scholar
20 Zhao, X., Shi, Y., Tang, J., Tang, R., Yu, L., Gu, N., Feng, G., Zhu, S., Liu, H., Xing, Y., et al. (2004) J. Med. Genet. 41, 31–34.Crossref, Google Scholar
21 Iwata, N., Suzuki, T., Ikeda, M., Kitajima, T., Yamanouchi, Y., Inada, T. & Ozaki, N. (2004) Mol. Psychiatry 9, 126–127.Crossref, Google Scholar
22 Thiselton, D. L., Webb, B. T., Neale, B. M., Ribble, R. C., O’Neill, F. A., Walsh, D., Riley, B. P. & Kendler, K. S. (2004) Mol. Psychiatry 9, 777–783.Crossref, Google Scholar
23 Gerlai, R., Pisacane, P. & Erickson, S. (2000) Behav. Brain Res. 109, 219–227.Crossref, Google Scholar
24 Bao, J., Wolpowitz, D., Role, L. W. & Talmage, D. A. (2003) J. Cell Biol. 161, 1133–1141.Crossref, Google Scholar
25 Rimer, M., Barrett, D. W., Maldonado, M. A., Vock, V. M. & Gonzalez-Lima, F. (2005) NeuroReport 16, 271–275.Crossref, Google Scholar
26 Lipska, B. K. (2004) J. Psychiatry Neurosci. 29, 282–286.Google Scholar
27 Hashimoto, R., Straub, R. E., Weickert, C. S., Hyde, T. M., Kleinman, J. E. & Weinberger, D. R. (2004) Mol. Psychiatry 9, 299–307.Crossref, Google Scholar
28 Farrer, M., Chan, P., Chen, R., Tan, L., Lincoln, S., Hernandez, D., Forno, L., Gwinn-Hardy, K., Petrucelli, L., Hussey, J., et al. (2001) Ann. Neurol. 50, 293–300.Crossref, Google Scholar
29 Bertram, L., Hiltunen, M., Parkinson, M., Ingelsson, M., Lange, C., Ramasamy, K., Mullin, K., Menon, R., Sampson, A. J., Hsiao, M. Y., et al. (2005) N. Engl. J. Med. 352, 884–894.Crossref, Google Scholar
30 Harrison, P. J. (2004) Psychopharmacology 174, 151–162.Crossref, Google Scholar
31 Law, A. J., Shannon, W. C., Hyde, T. M., Kleinman, J. E. & Harrison, P. J. (2004) Neuroscience 127, 125–136.Crossref, Google Scholar
32 Alberti, S., Krause, S. M., Kretz, O., Philippar, U., Lemberger, T., Casanova, E., Wiebel, F. F., Schwarz, H., Frotscher, M., Schutz, G., et al. (2005) Proc. Natl. Acad. Sci. USA 102, 6148–6153.Crossref, Google Scholar
33 Nagata-Ohashi, K., Ohta, Y., Goto, K., Chiba, S., Mori, R., Nishita, M., Ohashi, K., Kousaka, K., Iwamatsu, A., Niwa, R., et al. (2004) J. Cell Biol. 165, 465–471.Crossref, Google Scholar
34 Kleinjan, D. A. & van Heyningen, V. (2005) Am. J. Hum. Genet. 76, 8–32.Crossref, Google Scholar
35 Corfas, G., Roy, K. & Buxbaum, J. D. (2004) Nat. Neurosci. 7, 575–580.Crossref, Google Scholar
36 Thomas, J. O. & Travers, A. A. (2001) Trends Biochem. Sci. 26, 167–174.Crossref, Google Scholar
37 Cardon, L. R. & Abecasis, G. R. (2003) Trends Genet. 19, 135–140.Crossref, Google Scholar
38 Bejerano, G., Pheasant, M., Makunin, I., Stephen, S., Kent, W. J., Mattick, J. S. & Haussler, D. (2004) Science 304, 1321–1325.Crossref, Google Scholar
39 Hong, C. J., Huo, S. J., Liao, D. L., Lee, K., Wu, J. Y. & Tsai, S. J. (2004) Neurosci. Lett. 366, 158–161.Crossref, Google Scholar
40 Harrison, P. J. (1999). Brain 122, 593–624.Crossref, Google Scholar
41 Gu, Z., Jiang, Q., Fu, A. K., Ip, N. Y. & Yan, Z. (2005) J. Neurosci. 25, 4974–4984.Crossref, Google Scholar
42 Burbach, G. J., Dehn, D., Del Turco, D. & Deller, T. (2003) J. Neurosci. Methods 131, 83–91.Crossref, Google Scholar
43 Abecasis, G. R. & Cookson, W. O. (2000) Bioinformatics 16, 182–183. Abbreviations: htSNP, haplotype-tagging SNP; LD, linkage disequilibrium; NRG1, neuregulin 1; PMI, postmortem interval; SRF, serum response factor.Crossref, Google Scholar