Advances and Challenges in the Genetics of Autism
Abstract
Autism is characterized by (A) qualitative impairment in social interaction, (B) qualitative impairment in communication, and (C) repetitive behaviors and restricted interests (DSM-IV). As might be expected for a disorder with qualitative impairments in multiple domains, autism is extremely heterogeneous. In addition to variability in the triad of core symptoms, there is also heterogeneity in co-occurring medical conditions among children with autism. Adding further to the heterogeneity, there is also a broader group of diagnoses called the autism spectrum disorder (ASD). ASD includes autism, Asperger's syndrome, Rett syndrome, and pervasive developmental disorders not otherwise specified, which has evolved to encompass a wide range of social and emotional abnormalities with varying levels of cognitive functioning. [For a comprehensive review of the diagnostic criteria for ASD, see the recent FOCUS review of Westphal and Volkmar (1).]
Despite its widely varying clinical presentation, ASD appears to be highly heritable. The time-honored way to determine the degree to which genetics contributes to risk of a particular disorder is to compare the concordance rate of the disorder in monozygotic twins and dizygotic twins. Twins are assumed to share most of the environmental factors that might contribute to disorder risk. However, monozygotic twins are genetically 100% identical, whereas dizygotic twins share only as much genetic information as typical siblings. Therefore, if the concordance rate (the percentage of twins in which both individuals are affected by the disorder) is higher in monozygotic twins than in dizygotic twins, then there is good evidence that genetic factors contribute to the disorder. For ASD, six such studies have been reported, and each of them provides strong evidence that ASD is highly heritable (2–7). In the first five studies, there were relatively few twin pairs (21–50 pairs) available for study, but there was general agreement that concordance rates were 80%–95% in monozygotic twins and 0%–30% in dizygotic twins (2–6). The most recent report included 277 twin pairs, and found concordance rates of 88% in monozygotic twins and 31% in dizygotic twins (7). These concordance rates indicate that genetic factors have a larger impact on ASD than on any other psychiatric disorder.
Despite its high heritability, the utility of clinical testing for ASD is limited at the current time (8–11). Rare syndromic disorders with high rates of co-occurring ASD account for ∼10% of individuals with ASD (8, 12). A high-resolution karyotype may detect duplication of chromosome 15q11–13, which is strongly implicated in ASD risk (evidence discussed below). However, the chromosome 15q11–13 duplication is observed in only ∼1% in individuals with ASD, a rate that may not justify broad clinical testing. Similarly, clinical fragile X testing will identify the ∼1% of individuals in whom ASD is due to co-occurring fragile X syndrome (13, 14). Among ASD, only Rett syndrome is understood at a genetic level that justifies clinical testing, focused on female patients with ASD and intellectual disability (15, 16). However, even the single-gene Rett syndrome, which is caused by various mutations of the MECP2 gene, can be genetically complex (17). Recent advances in ASD genetics suggest that routine testing may soon be justified in a larger subset of individuals with ASD (9). This review summarizes that recent evidence.
THE PAST: A BRIEF HISTORY OF ASD GENETIC STUDIES
Given the highly heritable nature of ASD, it is not surprising that a great deal of effort has been exerted to determine its genetic causes. Genetic linkage studies have proven to be powerful tools to find rare genetic mutations with high impact in disorders. Linkage studies require genotyping of families with multiple affected individuals to identify genetic markers that are “identical by descent” and correlated with diagnosis of the disorder. The first genome-wide ASD linkage study indicated the most significant signal on chromosome 7q (18). The majority of subsequent genome-wide linkage studies also indicated an ASD linkage peak on chromosome 7q, although linkage evidence also emerged for a number of other genomic regions, including chromosomes 2q, 3p, 15q, and 17q (19–27). Collectively, the results of the genome-wide linkage studies suggested that there are multiple genetic loci that contribute to ASD risk. The linkage studies effectively narrowed the search for ASD candidate genes to a few chromosomal regions. However, each of the linkage peaks was broad enough to span dozens of genes. Therefore, candidate gene association studies were necessary to pinpoint the genes that contributed to the linkage peaks.
Candidate gene association studies for ASD have been conducted on at least 196 different genes, with at least one report of positive association for 106 different genes (28). However, the majority (83 of 106) of these candidate genes have been positively associated in only a single study (28). Because even well-designed single candidate gene studies can generate spurious results, single gene association studies should be considered “tentative knowledge” (29). Genes should be considered candidates to contribute to disorder risk only after replication of the genetic association in multiple samples (29). Using these criteria, five genes stand out as candidates for ASD risk because positive association has been reported in four or more samples (28). Each of these genes maps to a chromosomal region that was implicated in previous genome-wide linkage studies: OXTR on chromosome 3p; MET and EN2 on chromosome 7q; GABRB3 on chromosome 15q; and SLC6A4 on chromosome 17q. It is not yet clear whether these results represent a confirmation of the genome-wide linkage results or a bias in the selection of candidate genes. The preponderance of evidence in favor of a linkage peak on chromosome 7q led to intense investigation of candidate genes in this region (30–38).
THE PRESENT: PARADIGM SHIFTS IN TECHNOLOGY AND EXPERIMENTAL APPROACH APPLIED TO ASD
Technological advances in genetics have been applied to ASD at a remarkable pace. The first genome-wide linkage study of ASD, published in 1998, examined 354 microsatellite markers (18). Assays that genotype individual single nucleotide polymorphism (SNP) markers in hundreds of samples (39) were applied widely in ASD candidate gene studies by 2004 (40). Most recently, genome-wide association study (GWAS) platforms that genotype 10,000 or 500,000 SNPs have been applied to ASD (41, 42). The latest technologies for copy number variations (deletions or duplications of DNA segments) have also been applied to ASD (43–47). All of these genome-wide technologies offer hope of identifying genetic variants that contribute to ASD susceptibility.
The other paradigm shift in ASD has been the experimental approach toward increasingly larger sample sizes. Early reports of linkage and association for ASD included less than 200 families. Reports from other complex genetic disorders indicated that success depended on much larger sample sizes; reproducible genome-wide genetic associations were found in samples of thousands of patients (48, 49). Appropriately, the latest ASD genetics studies have included more than a thousand families (41, 42).
Applications of these genome-wide techniques to large sample sizes have begun to define chromosomal regions most likely to harbor genetic variants that contribute to ASD susceptibility. The genome-wide techniques have not pointed to the same chromosomal region (Figure 1). Initial attempts to replicate reports of genome-wide significant linkage, genome-wide significant association, and association of specific copy number variants (CNVs) with ASD risk have failed. However, these powerful genome-wide approaches have provided support for two previously identified ASD candidate genes and identified, for the first time, genetic loci with genome-wide significance that deserve further investigation.
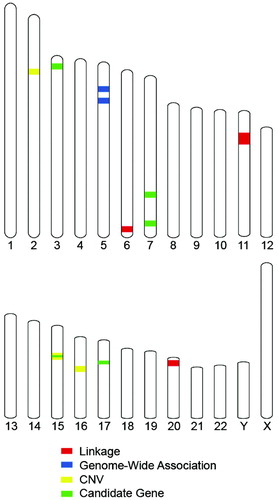
Figure 1. Summary of recent strong evidence for genetic contribution to ASD risk. The evidence for linkage, genome-wide association, recurrent CNV, and candidate gene association mapped to human chromosomes. The strongest evidence from each of these approaches points to multiple chromosomes, with convergence only on the GABRB3 gene on chromosome 15.
Linkage
Linkage analysis is a powerful technique to identify chromosomal regions that contribute to disease risk. Because the goal is to identify chromosomal regions that are coinherited with disease, linkage analysis requires families with multiple affected individuals. Ten ASD linkage analyses were published between 1998 and 2007 (18–27). Although a majority of studies identified linkage on chromosome 7q21–35, suggestive evidence for linkage has been described on every chromosome (50).
Two recent publications provided more robust evidence for linkage peaks in ASD. The Autism Genome Project Consortium performed linkage analysis on 10,000 SNP markers genotyped in 1,181 families (41). With this powerful combination of marker density and sample size, this group was able to identify a single linkage peak at chromosome 11p12–13 that exceeds the threshold for genome-wide suggestive linkage, particularly in families that have at least one female with ASD (41). Only modest support was observed for the previously identified linkage peaks on chromosomes 2q and 7q; support for a linkage peak on chromosome 17q was lacking (41). The results of this study suggest that rare genetic variants on chromosome 11p12–13 may contribute to ASD risk.
The most recent attempt at linkage analysis in ASD used technology that genotyped 500,000 SNPs in 878 families (42). Because marker densities that are too high can create statistical problems in linkage analyses, the authors pruned the number of markers used in their analyses to 16,311 highly polymorphic, high-quality SNPs (42). They found significant genome-wide linkage on chromosome 20p13 and suggestive evidence for linkage on chromosome 6q27 (42). These linkage data provided no evidence in support of the previously identified linkage peaks on chromosomes 2q, 7q, and 17q (42). Neither chromosome 20p13 nor chromosome 6q27 was suggestive for linkage in the Autism Genome Project Consortium study (41). Similarly, Weiss et al. (42) did not find evidence to support the Autism Genome Project Consortium linkage peak on chromosome 11p12–13 (41). Thus, the two large, recent linkage scans identified linkage signals different from each other and linkage signals different from those reported previously, confirming the heterogeneity of ASD. Resequencing and candidate gene association studies will be required to identify the functional variants contributing to each of the linkage signals.
GWAS
The advancement of technology platforms that genotype 10,000, 500,000, or even 1 million SNPs allows the study of genetic association on a genome-wide scale, instead of studying individual candidate genes. This new technology provides a powerful approach for identifying common genetic variants that contribute to ASD risk.
The first published GWAS of ASD found evidence of a genome-wide significant association signal on chromosome 5p14 (51), outside any previously reported linkage peak for ASD. The significant association signal lies between the genes encoding cadherin 9 (CDH9) and cadherin 10 (CDH10), both of which encode cell adhesion molecules that are expressed in the developing brain (51). The genetic evidence is extremely compelling and convincing: the original genome-wide significant association signal in 780 families was replicated in a large case-control sample. Further, the association peaked at a single SNP (rs4307059) that was flanked by several other markers that also showed evidence of association (51), suggesting that the association at rs4307059 was not due to a technical artifact. Because brain expression levels of CDH9 and CDH10 do not correlate with genotype at rs4307059 (51), the functional variant is yet to be identified.
The second published GWAS association data failed to replicate the association of rs4307059 on chromosome 5p14 (42). Instead, the second GWAS identified genome-wide significant association of markers ∼16 million base pairs away, on chromosome 5p15 (42). Here, three markers displayed genome-wide significant association near the gene encoding semaphorin 5A (SEMA5A). Like the cadherins, semaphorin 5A is a cell adhesion molecule with a defined role in brain development. Further, an extensive study of postmortem brains revealed a significant 20% decrease in SEMA5A transcript in postmortem brains of individuals with ASD compared with control subjects (42). It is not yet clear whether the identified genetic variants correlate with SEMA5A expression levels. Moreover, the identified risk allele for ASD near the SEMA5A gene is present at a frequency of >95% (42), suggesting that direct resequencing will be required to identify a less common, functional risk allele.
Collectively, the two reported ASD GWAS studies point to regions of chromosome 5p that may include common genetic variants that contribute to ASD risk. These studies provide strong statistical evidence for a potential functional variant on chromosome 5p, a region that was not revealed in any linkage study of ASD. Additional studies will be required to determine the functional genetic variants.
CNV
Most cells of the human body contain 23 pairs of chromosomes. However, humans can survive with regions of chromosomes that are present in only one copy (“deletion”) or in three or more copies (“duplication”). Deletions and duplications are collectively known as CNVs. CNVs are more common than was appreciated only 10 years ago, and technology continues to improve to identify increasingly smaller regions of CNVs. CNVs do not always cause disease. In fact, healthy control individuals carry an average of 11 CNVs, suggesting that even multiple CNVs are not pathological (52). Nevertheless, increasing evidence suggests that CNVs are a significant source of genetic variation and that specific CNVs may contribute to ASD risk.
There are multiple technologies used for detection of CNVs. [For a recent review of these CNV technologies and how they have been applied to ASD and schizophrenia, see Merikangas et al. (47).] There are two types of CNVs: inherited and de novo. Inherited CNVs, as the name implies, are passed from one generation to the next. In contrast, de novo CNVs are not inherited from a parent but are instead mutations that are first observed in the patient. De novo CNVs are rare and more likely to be pathological than inherited CNVs.
Ten studies have reported analysis of CNVs in ASD. Although improved technology may provide improved resolution of smaller CNVs, three apparently definitive conclusions can be drawn from these studies. First, the results of several studies establish that genome-wide inherited CNVs are as common in individuals with ASD as in control individuals or unaffected siblings, indicating that there is no increased burden of CNVs in ASD (41, 44, 46). Second, genome-wide de novo CNVs appear at a frequency of 7%–10% in simplex families (those with a single individual with ASD), 2%–3% in multiplex families (those with multiple individuals with ASD), and ∼1% of control individuals (43, 44). The higher rate of de novo CNVs in simplex families compared with that in multiplex families suggests that de novo events may contribute to ASD risk in this subset of families. Third, there does not appear to be a single CNV that accounts for a large fraction of cases of ASD or is diagnostic for ASD. However, CNV analysis could provide important clues about what genes and biological pathways contribute to ASD risk. In the data presented to date, it appears that CNVs of three chromosomal regions may be enriched in ASD: chromosome 2p16, chromosome 15q11–13, and chromosome 16p11.2.
De novo CNV of chromosome 2p16 was first described in a single family (41). Because of a lack of control samples in this study, it was difficult to determine whether any of these de novo events indicated an increased frequency of ASD (41). However, the de novo CNV of chromosome 2p16 involved deletion of the gene encoding the synaptic protein neurexin 1 (NRXN1). Two subsequent reports indicated de novo deletion of NRXN1 in ∼0.5% of individuals with ASD that was absent from control individuals, suggesting that deletion of NRXN1 may contribute to ASD risk in a small fraction of patients. De novo deletion of NRXN1 was observed in 9 of 1,771 (0.5%) individuals with ASD but in none of 2,989 control individuals (53) and in 10 of 2,195 (0.5%) individuals with ASD but none of 2,519 control individuals (46). However, the opposite has also been reported: an absence of de novo deletion of NRXN1 in 2,252 individuals with ASD or developmental delay, but the presence of the NRXN1 deletion in 5 of 2,814 (0.2%) control individuals (45). Therefore, deletion of NRXN1 may contribute to ASD risk in a small fraction of patients, but it is not diagnostic for ASD.
De novo CNVs of chromosome 16p11.2 were also originally described in a single family (43). A subsequent study revealed that a de novo CNV of chromosome 16p11.2 was present in 3 of 427 (∼1%) individuals with ASD (44). Simultaneously, a much larger study confirmed that a de novo CNV of chromosome 16p11.2 was present at a rate of ∼1% (15 of 1,740) in individuals with ASD and only 0.05% (12 of 23,502) of control individuals (45). The region of consistent chromosome 16p11.2 CNV includes ∼25 genes (44, 45). However, a recent report indicated a control sample frequency for chromosome 16p11.2 CNV of ∼0.2% (4 of 2,519) (46), confirming that the CNV of chromosome 16p11.2 is not diagnostic for ASD. Further, recent data also implicate CNV of the same chromosome 16p11.2 region in mental retardation and/or multiple congenital anomalies (54), suggesting that CNV of chromosome 16p11.2 may not be specific to ASD.
Duplication of chromosome 15q11–13 in individuals with ASD was first described nearly 20 years ago by cytogenetic analysis (55). This chromosomal region was implicated in multiple genome-wide linkage scans for ASD, and inheritance of a duplication of chromosome 15q11–13 from the mother (but not from the father) is associated with ASD (56). Deletion of chromosome 15q11–13 results in Prader-Willi syndrome (when inherited from the father) and Angelman syndrome (when inherited from the mother). The ASD candidate region includes eight genes (57) and may contribute to ASD risk via multiple genetic and epigenetic mechanisms (58). Each of the recent CNV studies of ASD has provided strong support for contribution of chromosome 15q11–13 duplication to ASD risk (41, 43–46, 53). The rate of chromosome 15q11–13 duplication (inherited or de novo) is ∼1% in ASD. One de novo duplication included only two genes, ATP10A and GABRB3, narrowing the search for ASD candidate genes (45). A subsequent report indicates association of a rare functional variant of GABRB3 with ASD risk (59), providing evidence that this gene may contribute to the often replicated association of chromosome 15q11–13 with ASD risk.
One caveat to note about the CNV analyses to date is that most have used the Autism Genetics Resource Exchange (AGRE) collection, which comprises ∼95% multiplex families. Given the higher rate of de novo CNVs in simplex families, it is possible that important de novo CNVs that contribute to ASD risk are yet to be reported. Collectively, CNV studies of ASD have identified rare CNVs targeting several chromosomes other than chromosomes 2p16, 16p11.2, and 15q11–13 (41, 43–46, 53). Each of these preliminary observations awaits replication. It appears clear that chromosome 15q11–13 duplication, either de novo or inherited from the mother, contributes to ASD risk.
Candidate genes
The strongest ASD candidate genes are those for which convergent evidence exists. Ideally, linkage analysis would implicate the chromosomal region, candidate gene association studies would describe replicated association of alleles, GWAS studies would implicate the gene, rare CNVs of the gene would be identified, and there would be some functional evidence that the gene is involved in ASD risk. No candidate gene meets all of these criteria. However, there are five genes that have convergent evidence for contributing to ASD risk. The evidence for each is detailed below and summarized in Table 1.
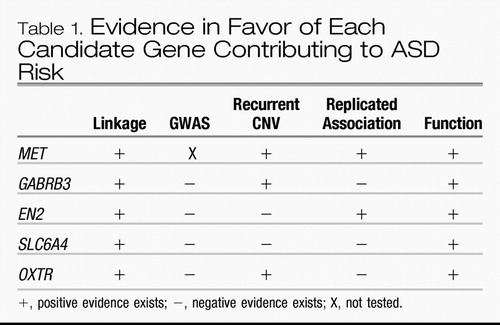 |
Table 1. Evidence in Favor of Each Candidate Gene Contributing to ASD Risk
MET.
The MET gene encodes the MET receptor tyrosine kinase, a key regulator of neuronal migration and synapse formation in the brain. Most linkage studies implicate the chromosome 7q31 region in which the MET gene lies. The results of all genetic association studies reported to date indicate positive association of MET gene variants. The MET promoter variant rs1858830 C allele was associated in five independent samples (37, 60, 61). Another study did not precisely replicate association of the MET rs1858830 C allele but instead found positive association of another genetic variant in MET that is likely to regulate its expression in two independent samples (62). The MET rs1858830 C allele is functional: it decreases transcription twofold due to altered binding of transcription factor complexes (37). Expression of MET protein is decreased twofold in postmortem brains of individuals with ASD (63). Further, the association of the MET rs1858830 C allele is enriched in individuals with co-occurring ASD and gastrointestinal conditions, indicating that MET may contribute to a subset of cases of ASD (64). Rare CNVs of the region including the MET gene have been reported (44). GWAS platforms lack markers near the ASD-associated MET promoter variant, and thus association of MET is not tested in GWAS studies (42). Therefore, overwhelming convergent evidence points toward association of common genetic variants in the MET gene that regulate its expression and contribute to ASD risk in a subset of patients with co-occurring gastrointestinal conditions.
GABRB3.
The GABRB3 gene encodes the GABAA receptor β3 subunit protein, a critical component of inhibitory signaling in the brain. Some, but not all, linkage studies of ASD implicated the chromosome 15q11–13 region in ASD. The results of many candidate gene association studies of GABRB3 were inconclusive. Association of the GABRB3 marker 155CA-2 was observed in two samples (65, 66) but not replicated in three other samples (67–69). Similarly, a positive association of the GABRB3 marker rs2081648 was observed in a study of 104 Korean trios, but this result was not replicated in 470 Caucasian families (70, 71). However, there is now overwhelming evidence in favor of chromosome 15q11–13 duplication contributing to ASD risk in ∼1% of families (41, 43–46, 53). Further, a functional rare variant, also present in ∼1% of families, is associated with ASD risk (59). The ASD-associated mutant form of the protein causes decreased expression of the receptor on the cell surface and reduced whole cell current (59). Therefore, despite the absence of a clear association of common genetic variants, accumulating evidence indicates that rare variation of the GABRB3 gene contributes to a subset of cases of ASD.
EN2.
The EN2 gene encodes ENGRAILED 2, a transcription factor involved in cerebellar development. Most linkage studies implicated the chromosome 7q36 location of the EN2 gene. Association of two alleles, the rs1861972 A allele and rs1861973 C allele, in the only intron of the EN2 gene was first observed in two samples (36) and then was replicated in four additional samples (72–74). A study of 210 Chinese Han families did not precisely replicate the association of rs1861972 and rs1861973 but instead found association of another SNP in the EN2 intron, rs3824068 (75). Another association analysis in a Chinese Han sample found association of the opposite alleles of rs1861972 and rs1861973 (76). Together, these results implicated the EN2 gene in ASD risk, but suggested that the functional variant had not been identified. However, recent functional studies indicate that the alleles of rs1861972 and rs1861973 bind different transcription factor complexes and cause a modest (∼20%) but significant change in transcriptional efficiency (77). Although recent CNV and GWAS data do not implicate EN2 and evidence of altered EN2 expression in ASD is lacking, association of common variants in EN2 in eight samples suggests that this gene contributes to at least a subset of cases of ASD.
SLC6A4.
The SLC6A4 gene encodes the serotonin transporter, a critical regulator of the neurotransmitter serotonin in both the brain and peripheral tissues. Because increased platelet serotonin is one of the few biomarkers that identify a subset of patients with ASD, the serotonin transporter is a plausible biological candidate for ASD risk. Multiple linkage studies found evidence for linkage of the chromosome 17q11.1–12 region at which the SLC6A4 gene resides. One of the first candidate gene association studies in ASD used microsatellite markers to describe association of the short allele of a SLC6A4 promoter variant (78). The short allele of the SLC6A4 promoter variant is functional: it decreases transcription efficiency, resulting in decreased gene expression and serotonin uptake activity (79). However, the results of more than a dozen genetic association studies in ASD have yielded mixed results: some reports found significant association of the short allele, some reports described significant association of the long allele, and some reports found no association of either allele (80). In fact, a recent meta-analysis of 14 studies indicates no overall association of the SLC6A4 promoter variant (81). Further, neither CNV nor GWAS studies implicate the SLC6A4 gene in ASD risk. However, rare functional variants may contribute to ASD risk, especially in a subset of patients with rigid-compulsive behaviors (82).
OXTR.
The OXTR gene encodes the oxytocin receptor, a known modulator of social behavior (83). Intranasal oxytocin administration was shown to improve the ability of individuals with ASD to recognize emotions (84), emphasizing the biological plausibility of OXTR contribution to ASD risk. One genome-wide linkage analysis highlighted the chromosome 3p24–26 region of OXTR (24). A study of 195 Chinese families indicated association of two markers in OXTR: rs2254298 A allele and rs53576 A allele (85). Association of the rs2254298 A allele was replicated in a Japanese case-control sample (86), and the opposite allele was identified in a study of 57 Caucasian families (87). Two additional studies failed to replicate association of rs2254298 with ASD (88, 89), but association of nearby markers in the OXTR gene have also been described (88, 90). In addition, a recent report described deletion of the region including OXTR and four neighboring genes in 1 of 119 families, altered methylation of the OXTR promoter in individuals with ASD, and decreased expression of OXTR in postmortem brains of individuals with ASD (91). Therefore, there may be multiple modes of disrupting OXTR that result in decreased oxytocin receptor and an increased risk for ASD.
Among the candidate genes with the most evidence in favor of contribution to ASD risk, there is little overlap with recent genome-wide linkage, GWAS, and CNV results (Figure 1). Association studies of recently implicated genetic regions should provide additional strong ASD candidate genes. One promising addition to the list is CNTNAP2, a gene that contributes to quantitative language impairments (92, 93) and may also contribute to ASD risk (38, 94). For all the candidate genes, it will be important to determine the biological pathways by which functional genetic variants contribute to ASD risk.
THE FUTURE: POTENTIAL KEYS TO UNDERSTANDING ASD GENETICS
Future technological advances will provide additional information about the genetic basis of ASD risk. For example, the application of next-generation sequencing techniques to ASD has not been reported. Next-generation sequencing has its limitations (95) but has the potential to provide important genome-wide information about common variants that are not tested on GWAS platforms, rare mutations, and rare CNVs that are enriched in individuals with ASD (96). Similarly, improved resolution of CNV platforms promises to identify increasingly smaller CNVs that may contribute to ASD risk. The application of these CNV platforms to simplex families, in which the burden of de novo CNV appears to be higher, may provide important insights into the genetic basis of ASD risk. Once these approaches identify candidate genes, the challenge then becomes translation of the genetic findings to determine the biological impact of the ASD-associated variants on brain development (12).
Beyond technological advances in genetics, there are two additional issues that must be addressed to understand the genetic basis of ASD risk. First, the observation that ASD affects four times as many males as females must be explained. There must be a genetic basis to this distinction, but no empirical data described to date explain it. Second, the absence of a single strong genetic signal indicates that multiple genes contribute to ASD risk, but there is debate about how these various genetic variants converge to result in ASD. One possibility is that there are genes that contribute independently to deficits in social interaction, language, and behavioral flexibility, and that ASD results from the convergence of multiple genetic risk factors for each of these independent domains. Alternatively, ASD may arise from a common biological mechanism (and thus the three ASD domains are inseparable), with the genetic complexity arising from disruption of multiple genes encoding proteins within the biological pathway (97). It will be important to make this distinction because it influences future biological approaches to understanding ASD. If ASD is the convergence of multiple genetic risk factors for each phenotypic domain, then it will be important to understand the biological interaction of proteins encoded by genes that influence the social domain (e.g., OXTR) and the language domain (e.g., CNTNAP2). On the other hand, if ASD results from disruptions of a common biological pathway, then it will be important to understand the multiple ways in which that pathway can be disrupted [e.g., the multiple genes encoding proteins of the MET signaling pathway (60)]. Network analysis of ASD candidate genes will provide important insights (98).
In summary, the recent application of powerful genome-wide technologies to large samples has implicated novel genetic regions in ASD risk. These findings also provide additional support for the candidate genes MET and GABRB3 (Figure 1). The available genetic evidence provides an opportunity to understand the biological basis of ASD. The challenges are now to identify functional variants, describe the contributions of the risk alleles to altered brain development, and determine effective treatment strategies based on biologically sound hypotheses.
1 Westphal A, Volkmar FR: An update on autism. FOCUS 2008; 6: 284– 292Link, Google Scholar
2 Folstein S, Rutter M: Genetic influences and infantile autism. Nature 1977; 265: 726– 728Crossref, Google Scholar
3 Ritvo ER, Freeman BJ, Mason-Brothers A, Mo A, Ritvo AM: Concordance for the syndrome of autism in 40 pairs of afflicted twins. Am J Psychiatry 1985; 142: 74– 77Crossref, Google Scholar
4 Bailey A, Le Couteur A, Gottesman I, Bolton P, Simonoff E, Yuzda E, Rutter M: Autism as a strongly genetic disorder: evidence from a British twin study. Psychol Med 1995; 25: 63– 77Crossref, Google Scholar
5 Le Couteur A, Bailey A, Goode S, Pickles A, Robertson S, Gottesman I, Rutter M: A broader phenotype of autism: the clinical spectrum in twins. J Child Psychol Psychiatry 1996; 37: 785– 801Crossref, Google Scholar
6 Taniai H, Nishiyama T, Miyachi T, Imaeda M, Sumi S: Genetic influences on the broad spectrum of autism: study of proband-ascertained twins. Am J Med Genet B Neuropsychiatr Genet 2008; 147B: 844– 849Crossref, Google Scholar
7 Rosenberg RE, Law JK, Yenokyan G, McGready J, Kaufmann WE, Law PA: Characteristics and concordance of autism spectrum disorders among 277 twin pairs. Arch Pediatr Adolesc Med 2009; 163: 907– 914Crossref, Google Scholar
8 Lintas C, Persico AM: Autistic phenotypes and genetic testing: state-of-the-art for the clinical geneticist. J Med Genet 2009; 46: 1– 8Crossref, Google Scholar
9 El-Fishawy P, State MW: The genetics of autism: key issues, recent findings, and clinical implications. Psychiatr Clin North Am 2010; 33: 83– 105Crossref, Google Scholar
10 Caglayan AO: Genetic causes of syndromic and non-syndromic autism. Dev Med Child Neurol 2010; 52: 130– 138Crossref, Google Scholar
11 Benvenuto A, Moavero R, Alessandrelli R, Manzi B, Curatolo P: Syndromic autism: causes and pathogenetic pathways. World J Pediatr 2009; 5: 169– 176Crossref, Google Scholar
12 Levitt P, Campbell DB: The genetic and neurobiologic compass points toward common signaling dysfunctions in autism spectrum disorders. J Clin Invest 2009; 119: 747– 754Crossref, Google Scholar
13 Garcia-Nonell C, Ratera ER, Harris S, Hessl D, Ono MY, Tartaglia N, Marvin E, Tassone F, Hagerman RJ: Secondary medical diagnosis in fragile X syndrome with and without autism spectrum disorder. Am J Med Genet A 2008; 146A: 1911– 1916Crossref, Google Scholar
14 Clifford S, Dissanayake C, Bui QM, Huggins R, Taylor AK, Loesch DZ: Autism spectrum phenotype in males and females with fragile X full mutation and premutation. J Autism Dev Disord 2007; 37: 738– 747Crossref, Google Scholar
15 Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY: Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet 1999; 23: 185– 188Crossref, Google Scholar
16 Chahrour M, Zoghbi HY: The story of Rett syndrome: from clinic to neurobiology. Neuron 2007; 56: 422– 437Crossref, Google Scholar
17 Renieri A, Meloni I, Longo I, Ariani F, Mari F, Pescucci C, Cambi F: Rett syndrome: the complex nature of a monogenic disease. J Mol Med 2003; 81: 346– 354Crossref, Google Scholar
18 A full genome screen for autism with evidence for linkage to a region on chromosome 7q. International Molecular Genetic Study of Autism Consortium. Hum Mol Genet 1998; 7: 571– 578Crossref, Google Scholar
19 Philippe A, Martinez M, Guilloud-Bataille M, Gillberg C, Rastam M, Sponheim E, Coleman M, Zappella M, Aschauer H, Van Maldergem L, Penet C, Feingold J, Brice A, Leboyer M: Genome-wide scan for autism susceptibility genes. Paris Autism Research International Sibpair Study. Hum Mol Genet 1999; 8: 805– 812Crossref, Google Scholar
20 Barrett S, Beck JC, Bernier R, Bisson E, Braun TA, Casavant TL, Childress D, Folstein SE, Garcia M, Gardiner MB, Gilman S, Haines JL, Hopkins K, Landa R, Meyer NH, Mullane JA, Nishimura DY, Palmer P, Piven J, Purdy J, Santangelo SL, Searby C, Sheffield V, Singleton J, Slager S, Struchen T, Vieland, B, Wang K, Svenson S: Winklosky B: An autosomal genomic screen for autism. Collaborative linkage study of autism. Am J Med Genet 1999; 88: 609– 615Crossref, Google Scholar
21 Risch N, Spiker D, Lotspeich L, Nouri N, Hinds D, Hallmayer J, Kalaydjieva L, McCague P, Dimiceli S, Pitts T, Nguyen L, Yang J, Harper C, Thorpe D, Vermeer S, Young H, Hebert J, Lin A, Ferguson J, Chiotti C, Wiese-Slater S, Rogers T, Salmon B, Nicholas P, Petersen PB, Pingree C, McMahon W, Wong DL, Cavalli-Sforza LL, Kraemer HC, Myers RM: A genomic screen of autism: evidence for a multilocus etiology. Am J Hum Genet 1999; 65: 493– 507Crossref, Google Scholar
22 International Molecular Genetic Studies of Autism Consortium: A genomewide screen for autism: strong evidence for linkage to chromosomes 2q, 7q, and 16p. Am J Hum Genet 2001; 69: 570– 581Crossref, Google Scholar
23 Yonan AL, Alarcon M, Cheng R, Magnusson PK, Spence SJ, Palmer AA, Grunn A, Juo SH, Terwilliger JD, Liu J, Cantor RM, Geschwind DH, Gilliam TC: A genomewide screen of 345 families for autism-susceptibility loci. Am J Hum Genet 2003; 73: 886– 897Crossref, Google Scholar
24 Ylisaukko-oja T, Alarcon M, Cantor RM, Auranen M, Vanhala R, Kempas E, von Wendt L, Jarvela I, Geschwind DH, Peltonen L: Search for autism loci by combined analysis of Autism Genetic Resource Exchange and Finnish families. Ann Neurol 2006; 59: 145– 155Crossref, Google Scholar
25 Molloy CA, Keddache M, Martin LJ: Evidence for linkage on 21q and 7q in a subset of autism characterized by developmental regression. Mol Psychiatry 2005; 10: 741– 746Crossref, Google Scholar
26 McCauley JL, Li C, Jiang L, Olson LM, Crockett G, Gainer K, Folstein SE, Haines JL, Sutcliffe JS: Genome-wide and ordered-subset linkage analyses provide support for autism loci on 17q and 19p with evidence of phenotypic and interlocus genetic correlates. BMC Med Genet 2005; 6: 1Crossref, Google Scholar
27 Lauritsen MB, Als TD, Dahl HA, Flint TJ, Wang AG, Vang M, Kruse TA, Ewald H, Mors O: A genome-wide search for alleles and haplotypes associated with autism and related pervasive developmental disorders on the Faroe Islands. Mol Psychiatry 2006; 11: 37– 46Crossref, Google Scholar
28 Banerjee-Basu S, Packer A: SFARI gene: an evolving database for the autism research community. Dis Model Mech 2010; 3: 133– 135Crossref, Google Scholar
29 Sullivan PF: Spurious genetic associations. Biol Psychiatry 2007; 61: 1121– 1126Crossref, Google Scholar
30 International Molecular Genetic Studies of Autism Consortium: Further characterization of the autism susceptibility locus AUTS1 on chromosome 7q. Hum Mol Genet 2001; 10: 973– 982Crossref, Google Scholar
31 Persico AM, Militerni R, Bravaccio C, Schneider C, Melmed R, Trillo S, Montecchi F, Palermo M, Pascucci T, Puglisi-Allegra S, Reichelt KL, Conciatori M, Keller F: No association between the 4g/5G polymorphism of the plasminogen activator inhibitor-1 gene promoter and autistic disorder. Psychiatr Genet 2001; 11: 99– 103Crossref, Google Scholar
32 Hutcheson HB, Bradford Y, Folstein SE, Gardiner MB, Santangelo SL, Sutcliffe JS, Haines JL: Defining the autism minimum candidate gene region on chromosome 7. Am J Med Genet B Neuropsychiatr Genet 2003; 117B: 90– 96Crossref, Google Scholar
33 Bonora E, Beyer KS, Lamb JA, Parr JR, Klauck SM, Benner A, Paolucci M, Abbott A, Ragoussis I, Poustka A, Bailey AJ, Monaco AP: Analysis of reelin as a candidate gene for autism. Mol Psychiatry 2003; 8: 885– 892Crossref, Google Scholar
34 Bonora E, Lamb JA, Barnby G, Sykes N, Moberly T, Beyer KS, Klauck SM, Poustka F, Bacchelli E, Blasi F, Maestrini E, Battaglia A, Haracopos D, Pedersen L, Isager T, Eriksen G, Viskum B, Sorensen EU, Brondum-Nielsen K, Cotterill R, Engeland H, Jonge M, Kemner C, Steggehuis K, Scherpenisse M, Rutter M, Bolton PF, Parr JR, Poustka A, Bailey AJ, Monaco AP: Mutation screening and association analysis of six candidate genes for autism on chromosome 7q. Eur J Hum Genet 2005; 13: 198– 207Crossref, Google Scholar
35 Persico AM, D'Agruma L, Maiorano N, Totaro A, Militerni R, Bravaccio C, Wassink TH, Schneider C, Melmed R, Trillo S, Montecchi F, Palermo M, Pascucci T, Puglisi-Allegra S, Reichelt KL, Conciatori M, Marino R, Quattrocchi CC, Baldi A, Zelante L, Gasparini P, Keller F: Reelin gene alleles and haplotypes as a factor predisposing to autistic disorder. Mol Psychiatry 2001; 6: 150– 159Crossref, Google Scholar
36 Gharani N, Benayed R, Mancuso V, Brzustowicz LM, Millonig JH: Association of the homeobox transcription factor, ENGRAILED 2, 3, with autism spectrum disorder. Mol Psychiatry 2004; 9: 474– 484Crossref, Google Scholar
37 Campbell DB, Sutcliffe JS, Ebert PJ, Militerni R, Bravaccio C, Trillo S, Elia M, Schneider C, Melmed R, Sacco R, Persico AM, Levitt P: A genetic variant that disrupts MET transcription is associated with autism. Proc Natl Acad Sci USA 2006; 103: 16834– 16839Crossref, Google Scholar
38 Arking DE, Cutler DJ, Brune CW, Teslovich TM, West K, Ikeda M, Rea A, Guy M, Lin S, Cook EH, Chakravarti A: A common genetic variant in the neurexin superfamily member CNTNAP2 increases familial risk of autism. Am J Hum Genet 2008; 82: 160– 164Crossref, Google Scholar
39 Livak KJ, Flood SJ, Marmaro J, Giusti W, Deetz K: Oligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic acid hybridization. PCR Methods Appl 1995; 4: 357– 362Crossref, Google Scholar
40 McCauley JL, Olson LM, Dowd M, Amin T, Steele A, Blakely RD, Folstein SE, Haines JL, Sutcliffe JS: Linkage and association analysis at the serotonin transporter (SLC6A4) locus in a rigid-compulsive subset of autism. Am J Med Genet B Neuropsychiatr Genet 2004; 127B: 104– 112Crossref, Google Scholar
41 Szatmari P, Paterson AD, Zwaigenbaum L, Roberts W, Brian J, Liu XQ, Vincent JB, Skaug JL, Thompson AP, Senman L, Feuk L, Qian C, Bryson SE, Jones MB, Marshall CR, Scherer SW, Vieland VJ, Bartlett C, Mangin LV, Goedken R, Segre A, Pericak-Vance MA, Cuccaro ML, Gilbert JR, Wright HH, Abramson RK, Betancur C, Bourgeron T, Gillberg C, Leboyer M, Buxbaum JD, Davis KL, Hollander E, Silverman JM, Hallmayer J, Lotspeich L, Sutcliffe JS, Haines JL, Folstein SE, Piven J, Wassink TH, Sheffield V, Geschwind DH, Bucan M, Brown WT, Cantor RM, Constantino JN, Gilliam TC, Herbert M, Lajonchere C, Ledbetter DH, Lese-Martin C, Miller J, Nelson S, Samango-Sprouse CA, Spence S, State M, Tanzi RE, Coon H, Dawson G, Devlin B, Estes A, Flodman P, Klei L, McMahon WM, Minshew N, Munson J, Korvatska E, Rodier PM, Schellenberg GD, Smith M, Spence MA, Stodgell C, Tepper PG, Wijsman EM, Yu CE, Roge B, Mantoulan C, Wittemeyer K, Poustka A, Felder B, Klauck SM, Schuster C, Poustka F, Bolte S, Feineis-Matthews S, Herbrecht E, Schmotzer G, Tsiantis J, Papanikolaou K, Maestrini E, Bacchelli E, Blasi F, Carone S, Toma C, Van Engeland H, de Jonge M, Kemner C, Koop F, Langemeijer M, Hijmans C, Staal WG, Baird G, Bolton PF, Rutter ML, Weisblatt E, Green J, Aldred C, Wilkinson JA, Pickles A, Le Couteur A, Berney T, McConachie H, Bailey AJ, Francis K, Honeyman G, Hutchinson A, Parr JR, Wallace S, Monaco AP, Barnby G, Kobayashi K, Lamb JA, Sousa I, Sykes N, Cook EH, Guter SJ, Leventhal BL, Salt J, Lord C, Corsello C, Hus V, Weeks DE, Volkmar F, Tauber M, Fombonne E, Shih A, Meyer KJ: Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet 2007; 39: 319– 328Crossref, Google Scholar
42 Weiss LA, Arking DE, Daly MJ, Chakravarti A: A genome-wide linkage and association scan reveals novel loci for autism. Nature 2009; 461: 802– 808Crossref, Google Scholar
43 Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, Yamrom B, Yoon S, Krasnitz A, Kendall J, Leotta A, Pai D, Zhang R, Lee YH, Hicks J, Spence SJ, Lee AT, Puura K, Lehtimaki T, Ledbetter D, Gregersen PK, Bregman J, Sutcliffe JS, Jobanputra V, Chung W, Warburton D, King MC, Skuse D, Geschwind DH, Gilliam TC, Ye K, Wigler M: Strong association of de novo copy number mutations with autism. Science 2007; 316: 445– 449Crossref, Google Scholar
44 Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, Skaug J, Shago M, Moessner R, Pinto D, Ren Y, Thiruvahindrapduram B, Fiebig A, Schreiber S, Friedman J, Ketelaars CE, Vos YJ, Ficicioglu C, Kirkpatrick S, Nicolson R, Sloman L, Summers A, Gibbons CA, Teebi A, Chitayat D, Weksberg R, Thompson A, Vardy C, Crosbie V, Luscombe S, Baatjes R, Zwaigenbaum L, Roberts W, Fernandez B, Szatmari P, Scherer SW: Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet 2008; 82: 477– 488Crossref, Google Scholar
45 Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, Fossdal R, Saemundsen E, Stefansson H, Ferreira MA, Green T, Platt OS, Ruderfer DM, Walsh CA, Altshuler D, Chakravarti A, Tanzi RE, Stefansson K, Santangelo SL, Gusella JF, Sklar P, Wu BL, Daly MJ: Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med 2008; 358: 667– 675Crossref, Google Scholar
46 Glessner JT, Wang K, Cai G, Korvatska O, Kim CE, Wood S, Zhang H, Estes A, Brune CW, Bradfield JP, Imielinski M, Frackelton EC, Reichert J, Crawford EL, Munson J, Sleiman PM, Chiavacci R, Annaiah K, Thomas K, Hou C, Glaberson W, Flory J, Otieno F, Garris M, Soorya L, Klei L, Piven J, Meyer KJ, Anagnostou E, Sakurai T, Game RM, Rudd DS, Zurawiecki D, McDougle CJ, Davis LK, Miller J, Posey DJ, Michaels S, Kolevzon A, Silverman JM, Bernier R, Levy SE, Schultz RT, Dawson G, Owley T, McMahon WM, Wassink TH, Sweeney JA, Nurnberger JI, Coon H, Sutcliffe JS, Minshew NJ, Grant SF, Bucan M, Cook EH, Buxbaum JD, Devlin B, Schellenberg GD, Hakonarson H: Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature 2009; 459: 569– 573Crossref, Google Scholar
47 Merikangas AK, Corvin AP, Gallagher L: Copy-number variants in neurodevelopmental disorders: promises and challenges. Trends Genet 2009; 25: 536– 544Crossref, Google Scholar
48 Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, Brant SR, Silverberg MS, Taylor KD, Barmada MM, Bitton A, Dassopoulos T, Datta LW, Green T, Griffiths AM, Kistner EO, Murtha MT, Regueiro MD, Rotter JI, Schumm LP, Steinhart AH, Targan SR, Xavier RJ, Libioulle C, Sandor C, Lathrop M, Belaiche J, Dewit O, Gut I, Heath S, Laukens D, Mni M, Rutgeerts P, Van Gossum A, Zelenika D, Franchimont D, Hugot JP, de Vos M, Vermeire S, Louis E, Cardon LR, Anderson CA, Drummond H, Nimmo E, Ahmad T, Prescott NJ, Onnie CM, Fisher SA, Marchini J, Ghori J, Bumpstead S, Gwilliam R, Tremelling M, Deloukas P, Mansfield J, Jewell D, Satsangi J, Mathew CG, Parkes M, Georges M, Daly MJ: Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat Genet 2008; 40: 955– 962Crossref, Google Scholar
49 Imielinski M, Baldassano RN, Griffiths A, Russell RK, Annese V, Dubinsky M, Kugathasan S, Bradfield JP, Walters TD, Sleiman P, Kim CE, Muise A, Wang K, Glessner JT, Saeed S, Zhang H, Frackelton EC, Hou C, Flory JH, Otieno G, Chiavacci RM, Grundmeier R, Castro M, Latiano A, Dallapiccola B, Stempak J, Abrams DJ, Taylor K, McGovern D, Silber G, Wrobel I, Quiros A, Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, Brant SR, Silverberg MS, Taylor KD, Barmuda MM, Bitton A, Dassopoulos T, Datta LW, Green T, Griffiths AM, Kistner EO, Murtha MT, Regueiro MD, Rotter JI, Schumm LP, Steinhart AH, Targan SR, Xavier RJ, Libioulle C, Sandor C, Lathrop M, Belaiche J, Dewit O, Gut I, Heath S, Laukens D, Mni M, Rutgeerts P, Van Gossum A, Zelenika D, Franchimont D, Hugot JP, de Vos M, Vermeire S, Louis E, Cardon LR, Anderson CA, Drummond H, Nimmo E, Ahmad T, Prescott NJ, Onnie CM, Fisher SA, Marchini J, Ghori J, Bumpstead S, Gwillam R, Tremelling M, Delukas P, Mansfield J, Jewell D, Satsangi J, Mathew CG, Parkes M, Georges M, Daly MJ, Heyman MB, Ferry GD, Kirschner B, Lee J, Essers J, Grand R, Stephens M, Levine A, Piccoli D, Van Limbergen J, Cucchiara S, Monos DS, Guthery SL, Denson L, Wilson DC, Grant SF, Daly M, Silverberg MS, Satsangi J, Hakonarson H: Common variants at five new loci associated with early-onset inflammatory bowel disease. Nat Genet 2009; 41: 1335– 1340Crossref, Google Scholar
50 Yang MS, Gill M: A review of gene linkage, association and expression studies in autism and an assessment of convergent evidence. Int J Dev Neurosci 2007; 25: 69– 85Crossref, Google Scholar
51 Wang K, Zhang H, Ma D, Bucan M, Glessner JT, Abrahams BS, Salyakina D, Imielinski M, Bradfield JP, Sleiman PM, Kim CE, Hou C, Frackelton E, Chiavacci R, Takahashi N, Sakurai T, Rappaport E, Lajonchere CM, Munson J, Estes A, Korvatska O, Piven J, Sonnenblick LI, Alvarez Retuerto AI, Herman EI, Dong H, Hutman T, Sigman M, Ozonoff S, Klin A, Owley T, Sweeney JA, Brune CW, Cantor RM, Bernier R, Gilbert JR, Cuccaro ML, McMahon WM, Miller J, State MW, Wassink TH, Coon H, Levy SE, Schultz RT, Nurnberger JI, Haines JL, Sutcliffe JS, Cook EH, Minshew NJ, Buxbaum JD, Dawson G, Grant SF, Geschwind DH, Pericak-Vance MA, Schellenberg GD, Hakonarson H: Common genetic variants on 5p14.1 associate with autism spectrum disorders. Nature 2009; 459: 528– 533Crossref, Google Scholar
52 Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, Maner S, Massa H, Walker M, Chi M, Navin N, Lucito R, Healy J, Hicks J, Ye K, Reiner A, Gilliam TC, Trask B, Patterson N, Zetterberg A, Wigler M: Large-scale copy number polymorphism in the human genome. Science 2004; 305: 525– 528Crossref, Google Scholar
53 Bucan M, Abrahams BS, Wang K, Glessner JT, Herman EI, Sonnenblick LI, Alvarez Retuerto AI, Imielinski M, Hadley D, Bradfield JP, Kim C, Gidaya NB, Lindquist I, Hutman T, Sigman M, Kustanovich V, Lajonchere CM, Singleton A, Kim J, Wassink TH, McMahon WM, Owley T, Sweeney JA, Coon H, Nurnberger JI, Li M, Cantor RM, Minshew NJ, Sutcliffe JS, Cook EH, Dawson G, Buxbaum JD, Grant SF, Schellenberg GD, Geschwind DH, Hakonarson H: Genome-wide analyses of exonic copy number variants in a family-based study point to novel autism susceptibility genes. PLoS Genet 2009; 5: e1000536Crossref, Google Scholar
54 Bijlsma EK, Gijsbers AC, Schuurs-Hoeijmakers JH, van Haeringen A, Fransen van de Putte DE, Anderlid BM, Lundin J, Lapunzina P, Perez Jurado LA, Delle Chiaie B, Loeys B, Menten B, Oostra A, Verhelst H, Amor DJ, Bruno DL, van Essen AJ, Hordijk R, Sikkema-Raddatz B, Verbruggen KT, Jongmans MC, Pfundt R, Reeser HM, Breuning MH, Ruivenkamp CA: Extending the phenotype of recurrent rearrangements of 16p11.2: deletions in mentally retarded patients without autism and in normal individuals. Eur J Med Genet 2009; 52: 77– 87Crossref, Google Scholar
55 Gillberg C, Steffenburg S, Wahlstrom J, Gillberg IC, Sjostedt A, Martinsson T, Liedgren S, Eeg-Olofsson O: Autism associated with marker chromosome. J Am Acad Child Adolesc Psychiatry 1991; 30: 489– 494Crossref, Google Scholar
56 Cook EH Jr, Lindgren V, Leventhal BL, Courchesne R, Lincoln A, Shulman C, Lord C, Courchesne E: Autism or atypical autism in maternally but not paternally derived proximal 15q duplication. Am J Hum Genet 1997; 60: 928– 934Google Scholar
57 Dykens EM, Sutcliffe JS, Levitt P: Autism and 15q11–q13 disorders: behavioral, genetic, and pathophysiological issues. Ment Retard Dev Disabil Res Rev 2004; 10: 284– 291Crossref, Google Scholar
58 Jiang YH, Sahoo T, Michaelis RC, Bercovich D, Bressler J, Kashork CD, Liu Q, Shaffer LG, Schroer RJ, Stockton DW, Spielman RS, Stevenson RE, Beaudet AL: A mixed epigenetic/genetic model for oligogenic inheritance of autism with a limited role for UBE3A. Am J Med Genet A 2004; 131: 1– 10Crossref, Google Scholar
59 Delahanty RJ, Kang JQ, Brune CW, Kistner EO, Courchesne E, Cox NJ, Cook EH Jr, Macdonald RL, Sutcliffe JS: Maternal transmission of a rare GABRB3 signal peptide variant is associated with autism. Mol Psychiatry 2009; doi: https://doi.org/10.1038/mp.2009.118Google Scholar
60 Campbell DB, Li C, Sutcliffe JS, Persico AM, Levitt P: Genetic evidence implicating multiple genes in the MET receptor tyrosine kinase pathway in autism spectrum disorder. Autism Res 2008; 1: 159– 168Crossref, Google Scholar
61 Jackson PB, Boccuto L, Skinner C, Collins JS, Neri G, Gurrieri F, Schwartz CE: Further evidence that the rs1858830 C variant in the promoter region of the MET gene is associated with autistic disorder. Autism Res 2009; 2: 232– 236Crossref, Google Scholar
62 Sousa I, Clark TG, Toma C, Kobayashi K, Choma M, Holt R, Sykes NH, Lamb JA, Bailey AJ, Battaglia A, Maestrini E, Monaco AP: MET and autism susceptibility: family and case-control studies. Eur J Hum Genet 2009; 17: 749– 758Crossref, Google Scholar
63 Campbell DB, D'Oronzio R, Garbett K, Ebert PJ, Mirnics K, Levitt P, Persico AM: Disruption of cerebral cortex MET signaling in autism spectrum disorder. Ann Neurol 2007; 62: 243– 250Crossref, Google Scholar
64 Campbell DB, Buie TM, Winter H, Bauman M, Sutcliffe JS, Perrin JM, Levitt P: Distinct genetic risk based on association of MET in families with co-occurring autism and gastrointestinal conditions. Pediatrics 2009; 123: 1018– 1024Crossref, Google Scholar
65 Cook EH Jr, Courchesne RY, Cox NJ, Lord C, Gonen D, Guter SJ, Lincoln A, Nix K, Haas R, Leventhal BL, Courchesne E: Linkage-disequilibrium mapping of autistic disorder, with 15q11–13 markers. Am J Hum Genet 1998; 62: 1077– 1083Crossref, Google Scholar
66 Buxbaum JD, Silverman JM, Smith CJ, Greenberg DA, Kilifarski M, Reichert J, Cook EH Jr, Fang Y, Song CY, Vitale R: Association between a GABRB3 polymorphism and autism. Mol Psychiatry 2002; 7: 311– 316Crossref, Google Scholar
67 Maestrini E, Lai C, Marlow A, Matthews N, Wallace S, Bailey A, Cook EH, Weeks DE, Monaco AP: Serotonin transporter (5-HTT) and γ-aminobutyric acid receptor subunit β3 (GABRB3) gene polymorphisms are not associated with autism in the IMGSA families. The International Molecular Genetic Study of Autism Consortium. Am J Med Genet 1999; 88: 492– 496Crossref, Google Scholar
68 Salmon B, Hallmayer J, Rogers T, Kalaydjieva L, Petersen PB, Nicholas P, Pingree C, McMahon W, Spiker D, Lotspeich L, Kraemer H, McCague P, Dimiceli S, Nouri N, Pitts T, Yang J, Hinds D, Myers RM, Risch N: Absence of linkage and linkage disequilibrium to chromosome 15q11–q13 markers in 139 multiplex families with autism. Am J Med Genet 1999; 88: 551– 556Crossref, Google Scholar
69 Martin ER, Menold MM, Wolpert CM, Bass MP, Donnelly SL, Ravan SA, Zimmerman A, Gilbert JR, Vance JM, Maddox LO, Wright HH, Abramson RK, DeLong GR, Cuccaro ML, Pericak-Vance MA: Analysis of linkage disequilibrium in γ-aminobutyric acid receptor subunit genes in autistic disorder. Am J Med Genet 2000; 96: 43– 48Crossref, Google Scholar
70 Ma DQ, Whitehead PL, Menold MM, Martin ER, Ashley-Koch AE, Mei H, Ritchie MD, Delong GR, Abramson RK, Wright HH, Cuccaro ML, Hussman JP, Gilbert JR, Pericak-Vance MA: Identification of significant association and gene-gene interaction of GABA receptor subunit genes in autism. Am J Hum Genet 2005; 77: 377– 388Crossref, Google Scholar
71 Ashley-Koch AE, Mei H, Jaworski J, Ma DQ, Ritchie MD, Menold MM, Delong GR, Abramson RK, Wright HH, Hussman JP, Cuccaro ML, Gilbert JR, Martin ER, Pericak-Vance MA: An analysis paradigm for investigating multi-locus effects in complex disease: examination of three GABA receptor subunit genes on 15q11–q13 as risk factors for autistic disorder. Ann Hum Genet 2006; 70: 281– 292Crossref, Google Scholar
72 Benayed R, Gharani N, Rossman I, Mancuso V, Lazar G, Kamdar S, Bruse SE, Tischfield S, Smith BJ, Zimmerman RA, Dicicco-Bloom E, Brzustowicz LM, Millonig JH: Support for the homeobox transcription factor gene ENGRAILED 2 as an autism spectrum disorder susceptibility locus. Am J Hum Genet 2005; 77: 851– 868Crossref, Google Scholar
73 Brune CW, Korvatska E, Allen-Brady K, Cook EH Jr, Dawson G, Devlin B, Estes A, Hennelly M, Hyman SL, McMahon WM, Munson J, Rodier PM, Schellenberg GD, Stodgell CJ, Coon H: Heterogeneous association between engrailed-2 and autism in the CPEA network. Am J Med Genet B Neuropsychiatr Genet 2008; 147B: 187– 193Crossref, Google Scholar
74 Sen B, Surindro Singh A, Sinha S, Chatterjee A, Ahmed S, Ghosh S, Usha R: Family-based studies indicate association of Engrailed 2 gene with autism in an Indian population. Genes Brain Behav 2009; doi: https://doi.org/10.1111/j.1601-183X.2009.00556.xGoogle Scholar
75 Wang L, Jia M, Yue W, Tang F, Qu M, Ruan Y, Lu T, Zhang H, Yan H, Liu J, Guo Y, Zhang J, Yang X, Zhang D: Association of the ENGRAILED 2 (EN2) gene with autism in Chinese Han population. Am J Med Genet B Neuropsychiatr Genet 2008; 147B: 434– 438Crossref, Google Scholar
76 Yang P, Lung FW, Jong YJ, Hsieh HY, Liang CL, Juo SH: Association of the homeobox transcription factor gene ENGRAILED 2 with autistic disorder in Chinese children. Neuropsychobiology 2008; 57: 3– 8Crossref, Google Scholar
77 Benayed R, Choi J, Matteson PG, Gharani N, Kamdar S, Brzustowicz LM, Millonig JH: Autism-associated haplotype affects the regulation of the homeobox gene, ENGRAILED 2. Biol Psychiatry 2009; 66: 911– 917Crossref, Google Scholar
78 Cook EH Jr, Courchesne R, Lord C, Cox NJ, Yan S, Lincoln A, Haas R, Courchesne E, Leventhal BL: Evidence of linkage between the serotonin transporter and autistic disorder. Mol Psychiatry 1997; 2: 247– 250Crossref, Google Scholar
79 Heils A, Teufel A, Petri S, Stober G, Riederer P, Bengel D, Lesch KP: Allelic variation of human serotonin transporter gene expression. J Neurochem 1996; 66: 2621– 2624Crossref, Google Scholar
80 Devlin B, Cook EH Jr, Coon H, Dawson G, Grigorenko EL, McMahon W, Minshew N, Pauls D, Smith M, Spence MA, Rodier PM, Stodgell C, Schellenberg GD: Autism and the serotonin transporter: the long and short of it. Mol Psychiatry 2005; 10: 1110– 1116Crossref, Google Scholar
81 Huang CH, Santangelo SL: Autism and serotonin transporter gene polymorphisms: a systematic review and meta-analysis. Am J Med Genet B Neuropsychiatr Genet 2008; 147B: 903– 913Crossref, Google Scholar
82 Sutcliffe JS, Delahanty RJ, Prasad HC, McCauley JL, Han Q, Jiang L, Li C, Folstein SE, Blakely RD: Allelic heterogeneity at the serotonin transporter locus (SLC6A4) confers susceptibility to autism and rigid-compulsive behaviors. Am J Hum Genet 2005; 77: 265– 279Crossref, Google Scholar
83 Ebstein RP, Israel S, Lerer E, Uzefovsky F, Shalev I, Gritsenko I, Riebold M, Salomon S, Yirmiya N: Arginine vasopressin and oxytocin modulate human social behavior. Ann NY Acad Sci 2009; 1167: 87– 102Crossref, Google Scholar
84 Guastella AJ, Einfeld SL, Gray KM, Rinehart NJ, Tonge BJ, Lambert TJ, Hickie IB: Intranasal oxytocin improves emotion recognition for youth with autism spectrum disorders. Biol Psychiatry 2010; 67: 692– 694Crossref, Google Scholar
85 Wu S, Jia M, Ruan Y, Liu J, Guo Y, Shuang M, Gong X, Zhang Y, Yang X, Zhang D: Positive association of the oxytocin receptor gene (OXTR) with autism in the Chinese Han population. Biol Psychiatry 2005; 58: 74– 77Crossref, Google Scholar
86 Liu X, Kawamura Y, Shimada T, Otowa T, Koishi S, Sugiyama T, Nishida H, Hashimoto O, Nakagami R, Tochigi M, Umekage T, Kano Y, Miyagawa T, Kato N, Tokunaga K, Sasaki T: Association of the oxytocin receptor (OXTR) gene polymorphisms with autism spectrum disorder (ASD) in the Japanese population. J Hum Genet 2010: 55: 137– 141Crossref, Google Scholar
87 Jacob S, Brune CW, Carter CS, Leventhal BL, Lord C, Cook EH Jr: Association of the oxytocin receptor gene (OXTR) in Caucasian children and adolescents with autism. Neurosci Lett 2007; 417: 6– 9Crossref, Google Scholar
88 Lerer E, Levi S, Salomon S, Darvasi A, Yirmiya N, Ebstein RP: Association between the oxytocin receptor (OXTR) gene and autism: relationship to Vineland Adaptive Behavior Scales and cognition. Mol Psychiatry 2008; 13: 980– 988Crossref, Google Scholar
89 Wermter AK, Kamp-Becker I, Hesse P, Schulte-Korne G, Strauch K, Remschmidt H: Evidence for the involvement of genetic variation in the oxytocin receptor gene (OXTR) in the etiology of autistic disorders on high-functioning level. Am J Med Genet B Neuropsychiatr Genet 2010; 153B: 629– 639Crossref, Google Scholar
90 Yrigollen CM, Han SS, Kochetkova A, Babitz T, Chang JT, Volkmar FR, Leckman JF, Grigorenko EL: Genes controlling affiliative behavior as candidate genes for autism. Biol Psychiatry 2008; 63: 911– 916Crossref, Google Scholar
91 Gregory SG, Connelly JJ, Towers AJ, Johnson J, Biscocho D, Markunas CA, Lintas C, Abramson RK, Wright HH, Ellis P, Langford CF, Worley G, Delong GR, Murphy SK, Cuccaro ML, Persico A, Pericak-Vance MA: Genomic and epigenetic evidence for oxytocin receptor deficiency in autism. BMC Med 2009; 7: 62Crossref, Google Scholar
92 Alarcon M, Abrahams BS, Stone JL, Duvall JA, Perederiy JV, Bomar JM, Sebat J, Wigler M, Martin CL, Ledbetter DH, Nelson SF, Cantor RM, Geschwind DH: Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene. Am J Hum Genet 2008; 82: 150– 159Crossref, Google Scholar
93 Vernes SC, Newbury DF, Abrahams BS, Winchester L, Nicod J, Groszer M, Alarcon M, Oliver PL, Davies KE, Geschwind DH, Monaco AP, Fisher SE: A functional genetic link between distinct developmental language disorders. N Engl J Med 2008; 359: 2337– 2345Crossref, Google Scholar
94 Bakkaloglu B, O'Roak BJ, Louvi A, Gupta AR, Abelson JF, Morgan TM, Chawarska K, Klin A, Ercan-Sencicek AG, Stillman AA, Tanriover G, Abrahams BS, Duvall JA, Robbins EM, Geschwind DH, Biederer T, Gunel M, Lifton RP, State MW: Molecular cytogenetic analysis and resequencing of contactin associated protein-like 2 in autism spectrum disorders. Am J Hum Genet 2008; 82: 165– 173Crossref, Google Scholar
95 Hert DG, Fredlake CP, Barron AE: Advantages and limitations of next-generation sequencing technologies: a comparison of electrophoresis and non-electrophoresis methods. Electrophoresis 2008; 29: 4618– 4626Crossref, Google Scholar
96 Alkan C, Kidd JM, Marques-Bonet T, Aksay G, Antonacci F, Hormozdiari F, Kitzman JO, Baker C, Malig M, Mutlu O, Sahinalp SC, Gibbs RA, Eichler EE: Personalized copy number and segmental duplication maps using next-generation sequencing. Nat Genet 2009; 41: 1061– 1067Crossref, Google Scholar
97 Campbell DB, Warren D, Sutcliffe JS, Lee EB, Levitt P: Association of MET with social and communication phenotypes in individuals with autism spectrum disorder. Am J Med Genet B Neuropsychiatr Genet 2010; 153B: 438– 446Crossref, Google Scholar
98 Bill BR, Geschwind DH: Genetic advances in autism: heterogeneity and convergence on shared pathways. Curr Opin Genet Dev 2009; 19: 271– 278Crossref, Google Scholar